Introduction
Genomic epidemiology combines classical epidemiological methods with genome sequence data to track and monitor both endemic pathogens and the emergence and spread of novel pathogens. In this tutorial, we will follow an outbreak through different phases and use phylogenetic methods to answer questions pertinent to understanding and controlling the outbreak.
Disclaimer: this outbreak is a fictional scenario based on simulated data. It is not intended to be predictive or relate to any real-world outbreak, and any similarities are coincidental. For details on how this outbreak was simulated see Simulation Notes.
To undertake this tutorial, you will need to download a number of software packages in a format that is compatible with your computer system (all three are available for Mac OS X, Windows and Linux/UNIX operating systems):
EXERCISE 1: A cluster of cases of unknown aetiology
Quick identification of causative pathogen
The below sequence is a contig ~18,000 bases long. A similar contig was found in each of the assemblies for the initial cases. We will perform a quick BLAST search to identify a likely causative agent.
>contig1|pathogenX
ACCAAACAAGGGAGAATATGGATACGTTACAATATATAACGTATTTTTAAAACTTAGGAACCAAGACAAACACTTTTTGTCTTGGTATTGGATCCTCAAGAAATATATCATCATGAGTGATATCTTTGAAAAGGCGGCGAGTTTTAGGAGTTATCAATCTAAGTTAGGGACAGATGGGAGGGCTAGTGCAGCAACTGCTACTTAGACAACCAAGATAAGGGTATTTGTACCAGCTACTAATAGTCCAGAGCTCAGATGGGAACTAACATTGTTTGCACTTGATGTGATTAGATCTCCGAGTGCTGCCGAGTCAATGAAAGTTGGAGCTGCTTTTACACGCATCTCTATGTATTCAGAGAGACCCGGGGCTCTCATTAGAAGTCTCCTCAATGACCCAGACATTGCAGCTGTAATAATTGATGTTGGATCAATGGTCAACGGAATACCAGTGATGGAGAGGAGAGGAGACAAGGCTCAGGAGGAGATGGAAGGCTTGATGAGAATCCTCAAAACTGCTCGGGACTGCAGCAAGGGAAAGACACCTTTTGTTGACAGGCGAACTTACGGCCTACGGATAACAGACATGAGCACCCTGGTCTCTGCACTTATCACCATCGAGGCCCAGATCTGGATACTGATCGCTAAAGCAGTTACAGCTCCCGACACTCCCGAGGAAAGTGAAACTAGAAGATTGGCTAAATACGTCCAAGAAAAGAGAGTCAATCCGTTCTTTGCTCTAACTCAGCAATGGCTAACAGAAATGAGGAATCTGCTCTCCCAGAGTCTATCAGTAAGGAAGTTTATGGTTGAGATCCTCATAGAAGTCAAGAAAGGAGGATCTGCTAAAGGCAGAGCAGTAGAAATAATCTCAGACATTGGAAACTATGTCGAGGAAACTGGTATGGCAGGATTCTTCGCAACCATCAGATTCGGGTTGGAGACAAGGTATCCAGCCCTTGCACTCAACGAATTCCTGAGTGACCTCAACACCATCACAAGCTTGATGCTACTCTACAGCGAAATTGGCCCAAGGGTCCCTGATATGGTGCTTCTTGAAGAATCAATTCAGACTAAGTTTGCCCCTGGAGGTTACCCATTATTGTGGAGCTTTGCCATGGGTGTGGCTACTACTATTGACAGGTCTATGGGGGCATTAAATATCAATCGTGGTTATCTTGAGCCTATGTATTTCAGACTTGGCCAAAAATCAGCACGTCACCATGCTGGAGGAATTGCTCAAAACATGGCACATAGACTGGGACTAAGTTCAGATCAAGTTGCAGAACTCGCTGCTGCAGTTCAGGAAACATCAGCAGGAAGGCAAGAGAGTAACGTTCAGGCTAGAGAGGCAAAATTTGCTGCAGGAGGTGTGCTCATTGGAGGCAGTGATCAAGATGTCGATGAAGAGGAAGAACCTATAGAATAGAGTGGCAGACAGTCAGTTACCTTCAAAAGGGAGATGAGTATTGCATCCCTTGCTGACAGTGTACCGAGCAGTTCTGCGAGCACATCCGGTGGGACCAGATTGACTAATTCATTACTAAACCTCAGATCAAGACTGGCTGCTAAAGCAGCAAAAGAAGCCGCCTCATCCAATGCAACAGATGATCCAGCAATCAGCAACAGAACTCAAGGGGAATCAGAGAAGAAGAATAATCAAGACCTCAAACCTACTCAAAATGACCTTGATTTCGTCAGAGCTGATGTGTGACGTCTATTTCCAATATTCTACAATATCCAAAAATCTTTTTATGGTACACTATCATAATACGACACTAAGGGATCAACCACCTCAAAGTTGCGACTCGTTTTAATTATATTAATCAAATGATACTCTTTTATGGGCAAACCGAAGAACCAATGTCTACACGTAAATTGAGCTTTGGTATTGCAATCTAATACTTGCTCAAAATCTTGAACTATTTGTGTAATTTCTATCATCATAGAGTTATAAAGTTTTTATTATATAAGTTGGTGCAGATCTTTGGACATGAATTACACACTACACTCTAATGAAGATAAAATTTTCATTACATATTTAAGGACTATTTCCTATCCTTTCAATGGTGCTTGGTTATGAAGGTTTCTTAATTTAAACAAGCTACTATCTTCGCACTGGAATATACAATACCTCTTACCTCATTTCTTACTTTAATATCATGTTATTTTTTTGATAAGTCACTTAACTTGACCAAGGTCTACCAGGCAATTCTCACACAAGTGAACTGCAGTCCCAACTTAGATTAAACATAATCATGCAAAATCACTATTTTGCACTACTAACTTATTAAGAAAGACTTAGGATCCAAGATATTTACTCTAGGATATCCTATTAGACTTAGCAGTCATTGGTTGAGGGTTCCCCTTACAAAACTCTAACCTTCACTCTAATAACAATTCATCCAATGGATAAATTGGAACTAGTCAATGATGGCCTCAATATTATTGACTTTATTCAGAAGAACCAAAAAGAAATACAGAAGACATACGGACGATCAAGCATTCAACAACCCAGCATCAAAGCCCGAACAAAAGCCTGGGAAGATTTGCTGCAGTGCACCAGTGGAGAATCTGAACAAGTTGAGGGGGGAATGTCTATGGATGATGGAGATGTTGAAAGAAGAAACGTGGAGGATCGATCCAGTACTTCTCCCACAGATGGAACTATTGGAAAGAGAGTGTCGAACACCCGTGACTGGGCAGAAGGTTCAGATGACATACAACTGGACCCAGTGGTTACAGACTTTGTATACCATGATCATGGAGGAGAATGTACCGGATATGGATTTACTTCAAGCCCTGAGAGAGGGTGGAGTAATTACACATCAGGAGCAAACAATGGGAATGTATGTCTTGTATCTGATGCAGAGATGCTGTCCTATGCTCTCGAAATTGCAGTTTCTAAAGAAGATCGGGAAACTGATCTAGTTCATCTTGAGAATAAACTATCTACTAAAGGACTGAAGCCCACAGCAGTACCGTTCACTCCGAGAAACCTGTCTGATCCTGCAAAAGACCCTCCTGTGATTGCTGAACACTACTACGGACTAGGAGTTAAAGAGCAAAACGTTGGCCCCCAGACTAGCAAAAATGTCAATTTGGACAGCATCAAATTGTACACATCAGATGACGAAGAGGCAGATCCGCTTGAATTCGAAGATGAGTTAGCAGGGAGCTCAAGTGAAGTGATACCCGGCAATACTCCTGAAGATGAAGAGCCTTCAAGTGTTGGCGGAAAACCCAATGAATCGATTGGACATACAATCGAAGGCCAATCAACCCGAGACAGCCTCCAAGCCAAGGGCAACAAACCAGCAGATGAACCAGGAGCAGGACCGAAAGATTAGGCCGTGAAGGAAGAACAACCCCAGAAGAGGCTACCTATGTTAGCTGAAGAATTTGAGTGCTCTGGATCGGAAGACCCAATCCTTCGGGAGCTGCTGAAGGAGAACTCACTCATAAATTGTCAACAAGGGAAAGATGCTCAGCCTCCATATCATTGGAGCATCGAGAGGTCAATAAGCCCGGATAAGACTGTGATCGCCAACGGTGCTGTGCAAACTGCTGACAGGCAAAGACCAGGAACTCCGATGCCAAAGTACCGAGGCATTCCTATTAAAAAGGGCACAGACGCGAAATATCCATCTGCTGGGACGGAAAACGTGCCTGGGTCGAAGAGTGGTGCAACCCGGCATGTTCGAGGATCACCCCCCTACCAAGAAGGCAAGAGTGTCAATGCGGAGAATGTCCAATTGAATGCTTCCACTGCGGTGAAGGAAACTGATAAGGCAGAAGTAAACCCCGCAGACGACAACGACTCACTCGATGATAAATACATCATGCCTTCAGATGATTTCTCAAACACTTTCTTCCCGCACGACACTGATCGCTTGAATTATCACGCAGATCATTTAGTTGATTATTACCTCGAAACCCTGTGTGAAGAGTCAGTTCTGATGGGAGTGATCAACTCTATAAAATTAATTAATCTGGACATGCGCTTAAATCACATTGAAGAACAAGTTAAAGAGATCCCAAAGATCATCAATAAGCTTGAGTCCATTGACAGAGTTTTGGCCAAGACCAATACCGCACTCTCAACCATTGAAGGACACCTGGTTTCCATGATGATAATGCTACCAGGGAAAGGGAAAGGAGAAAGAAAGGGCAAAAGTAATCCTGAGCTTAAACCAGTGATAGGAAGAGACATTCTAGAGCAGCAATCTCTTATCTCTTTTGACAATTTCAAGAATTTCAGAGATGGATCGTTGACAAACGAACCGTATGGGGCAGCTGTACAATTGAGAGAAGATCTTATTCTTCCTGAACTTAAGTTTGAGGAGACAAATGCATCTCAATTTGTTCCTATGGTAGATGGTTCATCCAGAGATGTTGTCAAGACATTGATAAGGACTCACATTAAAGATAGAGAGTTGAGATCAGAACTGATTGGTTACCTGAATAAAGCGGAAAATGATGAGGAAATTCAGGAGATAGCCAACACTGTCAATGACATCATTGACGGTAACATTTGATCACTGAATTGTCAGCAGAAATACAATGATCTATCAACAATCTCCCACAAGTAGACAATGGTTTCAGGTCAATAATAACAACCTCAATACTAATCTTTCACATAAGCATTACCCATTCTAGCCCTCAGACGATAACACAGTACTTGATACATGTTTATTGAAGTGTATGTAGCATGATTGAACTATTCAATCACTGTATTTCTCACTGTTGCTCTTAGTTAGTCATTGTATCTAATAATTATTATTACAGTACAAGGTATTATGAATTCAAAGATACGCAATAAATCTGATATCAGCATAGAGTAGAAAATTGTTGTTTTTGTCATGATCATTAGAAGATTTAACAATGATGTCAACTATCGTACCTAAACACAATAACATAAAATGGTCGATTTGTATTGTAGATCTCTCACGCATTTTAGTCTTATGAATTAGTGTTTCAAATCAGTTGCATATCAATTAAGAAAAACTTAGGAGACAGGTATAGAACCTCTCTTTCAGATAACTCGTCAATTAAGGACAGAAATTCTGTTTCTCAAATCCGCTAGCCTTTGCCAGAGAGGACACAAGCAATGTAGCCGGACATCAAGAGTATTTCAAGTGAGTCAATGGAAGGAGTATCTGATTTCAGCCCTAGTTCTTGGGAGCATGGTGGGTATCTTGATAAGGTTGAACCAGAAATTGATTAAAATGGCAGTATGATTTCAAAATACAAGACCTAAACCCCAGGAGCTAACGAGAGGAAATACAACAACTACATGTACCTTATATGTTACGGCTTTGTTGAAGATGTTGAGAGAACCCCAGAGACAGGGAAACGCAAGAAGATCAGGACAATTGCTGCCTCCCCTCTGGGTGTTGGTAAGAGTGCCTCTCATCCCCTAGATCTTCTGGAGGAACTCAGTTCCCTCAAAGTTACTGTGAGGAGAACAGCTGGATCAACTGAGAAAATTGTGTTTGGATCATCTGGCCCTCTAAATCACCTCGTTCCCTGGAAGAAAGTACTGACTGGTGGTTCAATTATTAATGCAGTCAAGGATTGTCGGAACGTTGATCAGATACAGCTCGACAAGCATCAAGCTCTGAGAATATTTATTCACAGTATCACAAAGCTCAATGATTCTGGAATCTACATGATTCCACGAACCATGCTAGAGTTCAGGAGAAACAATGCCATTGCCTTCAATCTTCTAGTGTACTTGAAGATTGATGCTGATTTATCCAAAATGGGGATCCGGGGAAGCCTCGATAAAGATGGCTTCAAGGTTGCCTCCTTCATGCTACACTTGGGGAAATTTGTCCGTCGTGCAGGGAAGTATTACTCTGTTGATTATTGAAGGAGAAAGATTGATAGGCTGAAATTGCAGTTTTCACTGGGTTCCATAGGCGGACTAAGTCTCCACATTAAGATCAATGGTGTAATCAGCAAACGGCTGTTTGCTCTAATGGGATTCCAAAAATACCTTTGTTTCTCCTTGATGGACATCAATCCTTGGCTCAACAGATTGACCTGGAACAACAGTTGTGAGATCAGCCGAGTAGCAGCTGTCTATCAGCCTTCTGTTCCAAGAGAGTTCATGATCTATGATGATGTCTTCATTGACAATACAGGGAGACTTCTAAAGGGCTAAACAGAATTCTTCTAAAATTTAATCAGTCATGAGTTTAGTAATCATACCTAGTCTTAATACATCACACAGGACTATTTACAAAAGACAATTAAAAAATAGAATAATCATGTAGTAGTAATTGAGAACATTATTAGAATGGTATAGCTAAAATGTAGTTTTTTTGAGTATTTGGTTTAAAATTAGATAACTATTACAAAAAACTTAGGAGCCAAGCTCTTGCCTCGTTCAGAAGTTTAAACAAGCATTCTTACCATTGGATAAACAAAAGGATTGGTTTTATCGTCTAAGAAATTTCTTGAAAGGCAAAGAGATTCCAGGTTTTATGTTGAATGAGGTCTATCAAACCAAGGAGACCCTCTAACAGCCAGGTCATAGGAATATAAATAAAAATAAGAATAAATATAAAATTGATTCCATCGAAAGATTCATTTCAAAAAGTGACCAAATAAAAGCGGTTGGCAGACCTACCAATCATATACCACAAGACTCGACAATGGTAGTTATACTTGACAAGAGATGTTAGTCTAATCTTTTAATACTGACTTTGATGATCTCGGAGTGTAGTGTTGGGCTTCTACATTATGAGAACTTGAGTAAAATTGGAATTGTCAAAGGAATAACAAGAAAATACAAGATTAAAAGCAATCCTCTCACAAAAGCCATTGTTATAAAAATGATTCCGAATGTGTCGAACATGTCTCAGTGCACAGGGAGTGTCATGGAAAATTATAAAACACGATTAAACGGTATCTTAACACCTATAAAGGGAGCGTTAGAGATCTACAAAAACAACACTCATGACCTTTTCGGTGATGTGAGATTAGCCGGAGTTTTAATGGCAGGAGTTGCTATTGGGATTGCAACCGCAGCTCAAATCACTGCAGGTGTAGCATTATATGAGGCAATGAAAAATGCTGACAACATCAACAAACTCTAAAGCAGCATTGAATCAACTAATGAAGCTGTCGTTAAACTTCAAGAGACTGCGGAAAAGACAGTCTATGTGCTGACTGCTCTACAGGATTACATTAATACTAACGTGGTACCGACAATTGACAAGATTAGCTGCAAACAGACAGAACTCTCACTTGATCTGGCATTATCAAAGTACCTTTCTGATTTGCTTTTTGTATTTGGCCCCAACCTTCAAGACCCAGTCTTTAAATCAATGACTATACAGGCTATATCTCAGGCATTCGGTGGAAATTATGAAACACTGCTAAGAACATTGGGTAACGCTACAGAAGACTTTGATGATGTTCTAGAAAGTGACAGCATAACGGGTCAAATCATCTATCTTGATCTAAGTAGTTAATATATAATTGTCAGGGTTTATTTTCCTATTCTCACTGAAATTCAACAGGCCTATATCCAAGAGTTGTTACCAGTGAGCTTCAACAATGATATTTCAGAATGGATCAGTATTGTCCCAAATTTCACATTGGTAAGGAATACATTAATATCAAATATAGAGATTGGATTTTGCCTAATTTCAAAGAGGAGCGTGATCTGCAACCAAGATTATGCCACACCTATGACCAACAACATGAGAGAATGTTCGACGGGATCGACTGAGAAGTGTCCTCGAGAGCTGGTTGTTTCATCACATGTTCCCAGATTTTCACTATCTAACGGGGTTCTGTTTGCCAATTGCATAAGTGTCACATGCCAGTGTCAAACAACAGGCAGGGCAATCTCACGGTCAGGAGAACAAACTCTGCTGATGATTAACAACACCACCTGTCCTACAGCCGTACTCGGTAATGTGATTATCAGCTTAGGGAAATATCTGGGGTCAATAAATTATAATTCTGAAGGCATTGCTTTCCGTCCTGCAGTCTTTACAGATAAAGTTGATATATCAAGTCAGATATCCAGCATGAATCAGTCCTTACAACAGTCTAAGGACTATCTCAAAGAGGCTCAACGACTCCTTGATACTGTTAATCCATCATTAATAAGCACGTTGTCTATGATCATAGTGTATGTACTATCGATCGCATCGTTGTGTATAGGGTTGATTACATTTATCAGTTATATCATTGTTAAGAAAAATAGAAACACCTACAGCAGATTAGAGGATAGAAGAGTCAGACCTACAAGCAGTGGGGATCTCTATTACATTGGGACATAGTGTATTCAGATTGATGAAATTATGTCAAAGAAATCAGAGAACTTCTGACTTTCAGAAATGGATTGTAGACAATTAGTTAGATCATCCTGAACAATCGAGGTGAAAACATTGCAACTTTAGAATCAGATCATGTAAATAGTTGTAAAAAATTATAAGCTTCTTTTAATTTGTTTGAACAATAATTTGATTAATATATAACATATTCGCTCACACGAGCGCTAACCTATACACTCTTTACTAATATGTTATACTCATAATTAATGATATAATGACAAATAAGGATTCAAATTGAATTATGATATAGTTTCACACTACAATAGCATTTCGACCCAGAAAATATCCTTACAATTATACAATGTACTTAACCGTGAATATGTAGTTGATAATTTCCCTTTAAAAATTTAATAAAAAACTTAGGACCCAGGTCCATAACTCATTGGATACTTAACTGTATACTTCTAAGCTATCACATATCAAAGGAGAGATTGAATGTTTTTTTAGAGATCTGGATCATTACTATATGTGTCTCCTATAATCACATCATAGGAGTCAGCCATAATACACATCTTTGGGTAAGGAAAGGAAAGTATTGTTGACGTACAGATTGATCTGCTTGAATCAAATAATCAGTCATAACAATTCAAGAGAATGCCGGCAGAAAGCAAGAAAGTTAGATCGGAAAATACTACTTCAGACAAAGGGAAATTTCCTAGTAAAATTATTAAGAGCTACTACGGTACCATGGACATTAACAAAATAAATGAAGGATTATTGGACAGCAAAATATTAAGTGCTTTCAACACAGTAATAGCATTGCTTGGATGTATCGTGATCATAGTGATGAATATAATGATCATCCAAAATTACACAAGATCAACAGACAATCAGGCCGTGATCAAACATGCGTTGCAGTGTATCCAACAGCAGATCAAAGGGCTTGCTGACAAAATCGGCACAGAGATAGGGCCCAAAGTATCACTGATTGACACATCCAGTACTATTACTATCCCAGCTAACATTGGGCTGTTAGGTTCAAAAATCAGCCAGTCGACTGCAAGTAGAAATGAGAATGTGAATGAAACATGCAAATTCACACTGCCTCCCTTGAAAATCCACGAATGTAACATTTCTTGTCGTAACCCACTCCCTTTTAGAGAGTATAGGCCGCAGACAGAAGGAGTGAGCAATCTGGTAGGATTACCTAATAATATTTGCCTGGAAAAGACATCTAATCAGATACTGAAGCCAAAGCTGATTTCATACACTTTACCCGTAGTCGGTCAAAGTGGTACCTGTATCCCAGACCCATTGCTGGCTATGGACGAGGGCTATTTTGCATATAGCCACCTGGAAAGAATCGGATCATGTTCAAGAGGGGTCTCCAAACAAAGAATAATAGGAGTTGGAGAGGTACTAGACAGAGGTGATGAAGTTCCTTCTTTATTTATGACCAATGTCTGGACCCCACCAAATCCAAACACCGTTTACCACTGTAGTCCTGTATACAACAATGAATTCTATTATGGGCTTTGTGCAGTGTCAACTGTTGGAGACCCTATTCTGAATAGCACTTACTGGTCCGGATCTCTAATGATGACCCGTCTAGCTGTAAAACCCAAGAGTAATGATGGGGGTTACAATCAACATCAACTTGCCCTACGAAGTATCGAGAAAGGGAGGTATGATAAAGTTATGCCGTATGGACCTTCAGGCATCAAACAGGGTGACACCCTGTATTTTCCTGCTGTAGGATTTTTGGTCAGGACAGAGTTCAAATACAATGATTCAAATTGTCCCATCACGAAGTGTCAATACAGTAAACCTTAAAATTACAGGCTTTCTATCGGGATTAGACCAAACAGCCATTATATCCTTCGATCTGGACTATTAAGATACAATCTATCAGATGGGGAGAACCCTAAAATTGTATTCATTGAAATATCTGATCAAAGATTATCTATTGGATCTCCTAGCAAAATCTCTGATTCTTTGGGTCAACCTGTTTTCTACCAAGCGTCATTTTCATGGGATACTATGATTAAATTTGGAGATGGTCAAACCGTCAACCCTATGGTTGTATATTGGCGTGATAAGACGGTAATATCAAGACCTGGGCAATCACAATGCCCTAGATTCAATACATGTCCAGAGATCTGCTGGGAAGGAGTTTATAATGATGCATTCCTAATTGACAGAATCAATTGGATAAGCGCGGGTGTACTCCTAGACAGCAATCAGACCGCAGAAAATCCTGTTTTTACTGTATTCAAAGATAATGAAATACTTAATAGAGCACAACTGGCTCCTAAGGACACCAATGCACAAAAAACAATAACTAATTGCTTTTTCTTGAAGAATAAGATTTGGTGCATATCATTGGTTGAGATATATGACACAGGAGACAATGTCATAAGACCCAAACTATTCGCAGCTACGATACCAGAGCAATGTACATAAAAATCAACCTCATAATTTAATGGATTGATCTAATATAATGATAATAACCGTACAAAGACATGTGATGTAAACAAAATTGTCGTAATTAAATAAGTCCTCAGCTGAATATTTTTTTAAGATTAGCAATAGCATGTTTATCCAGTTATTGGATAGTTGATAATTTAATTCTGAAACTGGGTTAATAAATAATCTTGATCGATGATCTTTGAGAACAATGATATCATATAGTTCATCAAGTGATAATCAATTCTTTATATGTACACTTTAGAGTATATTTTGAGACTTAGTATTTTCGGCCCGAATGTTAAAGTTAATAGTTCATACATAACCTAATCTCAAGTTCTAAGCATAATGATAACTATTAATGCGAACATGTCTTGATGTAAGGAAGATTTGATATCAACTGAGACTCCACTTGATATAGTAGAGCTGAATCTTGTAAATAAATTATAATGAATAGTTTATTCAAAGATTATCATTCATATCAGTATAATTTAAGAAAAACTTAGGACCCAGGTCCTTGATTGTGCCAATTTTCTTGAGAAATCATTCAATTGTCCTTAGACTGAAAGCGTTGTTACCTAGTTTTTCAGAAGAGATCTTATTAGAATTGATTTATATGATCTAATTCCCTTAAAAATTGAATACCAAAAAACAAAAATGGCCGATGAATTATCAATATCCGACATCATCTACCCTGATTGTCATTTGGATAGTCCTATAGTCTCTGGTAAACTAATATCAGCTATTGAATATGCTCAATTGAGACACAATCAACCCAGTGATGATAAAAGACTGTCTGAGAATATTAGGTTAAACCTTCACGGGAAAAGAAAGAGTCTATACATATTAAGACAATCCAAACAGGGTGATTACATTAGAAACAACATTAAAAACCTAAAGGAATTCCTGCATATTGCGTACCCTGAATGCGATTACACTTTATTCTCCATCACATCCCAAGGCATGACTAGCAAACTTGATAACATCATGCAAAAGTCATTCAAAGCATACAATATCATTAGTAAGAAAGTAATTGGGATGCTGCAAAATATCACTAGAAATCTCATAACTCAAGATAGAAGAGATGAAATAATTAATATACATGAGTGTAGGCGATTAGGGGATTTAGGGAAGAATATGAGTCAATCTAAATGGTATGAGTGTTTTTTGTTTTGGTTTACTATCAAAACAGAGATGCGAGCAGTGATCAAGAATTCGCCAAAGCCGAAATTCCGTTCAGATTCATGCATAATACACATGCGAGATAAAAGTACTCAAATAATCCTAAATCCAAATCTTATCTGCATTTTCAAATCAGACAAAACTGGATAGAAGTGTTATTATCTTACAACCGAAATGGTTCTAATGTATTGTGACGTCCTAGAGGGAAGGATGATGATGGAGACAACAGTCAAATCGGATATCAAGTACCAGCCTCTAATCTCGAGATCCAATGCCCTCTGGGGGCTAATGGATCCCTTGTTCCCTGTCATGGGAAACAGGATTTACAATATAGTGTCTATGATAGAGCCTTTAGTTCTTGCACTACTCCAACTCACGGATGAGGCGAGGATCCTGAGGGGTGCATTCCTGCATCACTGCATAAAGGAAATTCATCAAGAATTGAGTGAGTGTGGTTTTACAGATCAGAAGATTCGGTCTATGTTTATTGATGATCTTTTATCCATTCTAAATATCGATAATATACATCTGTTGGCAGAGTTCTTTTCTTTCTTTCGTACGTTTGGCCATCCTATTCTTGAGGCTAAAGTTGCTGCAGAAAAAGTGAGTGAACATATGTTGGCAGATAAAGTTCTTGAATATGCCCCTATAATGAAAGCACATGCTATATTCTGCGGGACTATAATAAATGGGTATAGGGATAGACACGGAGGAGCCTGACCTCCTCTTTACCTCCCCGCACATGCATCTAAGCATATAATCCGTTTGAAAAATTCTGGGGAATCTTTGACCATTGATGACTGTGTCAAGAATTGGGAATCATTCTGTGGGATTCAATTTGATTGTTTCATGGAGCTGAAATTGGACAGAGATCTGAGTATGTATATGAAAGCTAAAGCTTTATTTCCAATCAAAGACGAATGGGACAGTGTATACCCACGTGAAGTGTTGAGCTATACCCCACCGAGGTCAACCCAGCCAAGAAGATTGGTTGACGTTTTTGTAAATGATGAAATCTTTGATCCATACAACATGCTAGAATATGTCTTATCCGGTGCTTATCTCGAGGATGAACAATTCAATGTTTCTTATAGCTTGAAGGAGAAAGAGACGAAGCAAGCTGGACGATTGTTCGCAAAGATGACCTACAAAATGCGTACATGTCAAGTCATAGCAGAGGCCCTGATAGCCTCAGGTGTCGGTAAATATTTTAAGGAGAACGGGATGGTTAAGGATGAGCACGAACTTTTGAAGACACTCTTCCAATTGTCTATTTCCTCAGTTCCTCGAGGGAACAGTCAGGGTAATGATCCTCAATCCATCAATAATATAGAAAGAGATTGCCAATACTTTAAAGGGGTCACCACCACTGTGAAAGACAATAAGAATAACTCTTTTAATAAGGTTAAATCTGCTCTCAATAATCCGTGCCAAGCTGACGGAGTCCATCATAACATGTCACCCAATACACGAGATCGTTATAAGTGTAGTAATACAAGTAAGTCTTTTCTCGATTATCATACCGAGTTTAATCCTCACAATCACTACAAATCAGACAATACAGAGGCGGCCGTACTGTCCAGGTATGAAGACAACACCGGGACAAAATTTGATACAGTAAGTGCATTTCTTACAACTGATCTTAAGAAATTCTGTCTCAATTGGAGATACGAATCAATGGCTATATTTGCTGAACGTCTGGATGAGATATACGGTTTACCTGGATTATTTAATTGGATGCACAAACGACTAGAAAGATCTGTTATCTATGTTGCAGACCCTAATTGCCCCCGTAATATTGACAAACATATGGAACTAGAAGAAACTCCTGAAGATGATATATTCATTCATTATCCTAAATGCGGTATTGAAGGATATAGCCAAAAAACATGGACTATAGCAACTATCCCCTTTTTATTCTTGAGTGCCTGTGCGACGAACACGAGGATTGCTGCAATTGTCCAAGGAGACAATGAATCAATTGCTATCACTCAAAAAGTTCATCCTAATCTTCCCTACAAGGTAAAGATAGAGATCTGTGCAAAGCAAGCTCAGCTGTATTTTGAAAGGTTGAGGATGAACTTAAGAGCCCTCGGCCACAATCTTAAAGCTACAGAATCTATCATCAGTACACATCTTTTTGTTTATTCGAAGAAAATTCATTATGATGGAGCTGTGCTGTCTCAGGCACTCAAATCAATGTCCAGATGTTGCTTTTGGTCAGAGACCCTGGTGGATGAAACTAGATCAGCTTGTAGTAACATCAGCACTACGATAGCTAAAGCTATAGAAAATGGGTTGTCAAGAAATGACGGCTATTGTATCAATATTTTGAAAGTAATTCAGCAGCTTCTCATATCAACTGAGTTTAGTATTAACGAGACATTCACACTGGATGTGACATCTCCCATTTCAAAGAATTTAGATTGGCTTATAACAGCTGCATTAATCCCGGCACCTATTGGAGGATTCAATTACCTTAATTTGTCTAGAATTTTTGTTAGAAATATAGGTGATCCGGTTACAGCATCTTTGGCTGATCTTAAAAGAATGATTGACCACAGTATTATGACTGAAAGCGTATTACAAAAAGTTATGAATCGAGAACCAGGTGATGCGAGTTTCTTGGACTGGGCCAGTGATCCATACTCGGGCAACTTGCCTGACTCACAAAGCATCACTAAAACAATTAAAAATATCACAGCTAGGACTATACTGAGGAACTCACCGAATCCAATGCTAAAAGTTTTATTTTATGACAAATCTTTTGATGAAGATCTTGAACTAGCTAGCTTGTTAATGGACAGGAGGGTTATATTACCTAGAGCCGCTCATGAGATACTGGATAATTCATTGACAGGCGCCAGAGAGGAAATTGCTGGTTTATTAGATACAACTAAAGGCTTGATCAGATCAGGGCTAAGAAAGAGTCGAATTCAGCCAAAGTTAGTTTCTAGACTATCTCATCATGATTATAATCAATTTTTAAGACTGTATAAACTTCTATCAAACAGAAGACAAAATGACTTGATATCATCAAATACTTGCTCAGTTGACTTGGCACGAGCATTGAGATCTCACATGTGGAGCGAATTAGCTTTAGGTAGAGTAATATACGGACTTGAGGTTCCAGATGCACTTGAGGCTATGGTGGGAAGGTACATAACAGGGAGCTTAGAGTGCCAAATTTGTGAGCAGGGAAACACGATGCATGGGTGGTTCTTTGTACCTAGGGATTCCCAATTAGATCAGGTAGATAGAGAGCTCTCATCAATAAGAGTACCTTATGTATGATCAAGTACGGATGAAAGATCGGATATCAAACTAGGCAATGTCAAAAGACCAACTAAGGCCTTGCGTTCTGCTATCAGAATTGCGACAGTATATACTTGGGCCTATGGAGATAATGAAGAGTGTTGGTATGAAGCTTGGTACCTAGCGTCTCAGAGGGTAAACATAGACTTAGATGTATTGAAAGCTATAACCCCAGTATCCACTTCAAACAATTTATCCCATAGATTGAGAGATAAATCCACACAATTTAAGTTTGCAGGGAGTGAACTCAACAGGGTTTCTAGATATGTTAACATAAGCAATCATAGTCTAGATTTCAGAATTGAGGGGGAAAAGGTAGATACGAATCTTATTTATCAACAAGCAATGCTATTAGGGTTATCGCTATTGGAAGGTAAATTCAGATTGAGATTAGAAACTGATGATTACAACGGGATATATCACTTACATGTAAAGGATAATTGTTGTGTCAAAGAAGTGGCTGATGTAGGCCAAGTGGACGCTGAGTAGCCTATCCCAGAATATACTGAAGTGGATAACAATCATCTTATATATGATCCAGACCCCGTTACAGAAATTGATTGCAGCCGTCTTTCTAATCAGGAGTCCAAATCAAGAGAATTAGACTTTCCTTTATGGTCAACTGAGGAACTTCATGATGTCCTAGCTAATACTGTTGCTTAGACCGTTCATGAGATTATAACAAAGGCTGACAAGGATGTTTTAAAGCAACACCTTGCAATAGACTCTGACGAGAACATCAATAGCTTAATCACAGAATTTCTAATACTTGATCCTGAACTGTTTGCACTTTATCTAGGACAATCTATATCAATAAAATGGGCCTTTGAAATTCATCATAGGCGTCCTAGAGGAAGACATACTATGGTCGACCTATTGTCAGATCTTGTATCAAATACATCAAAACACACTTACAAAGTGTTGTCAAATGCCTTGTCACATCCTAGAGTATTCAAGAGATTTGTAAACTGTGGCTTACTATTGCCTACACAGGGTCCTTACCTTCATCAACAAGATTTTGAAAAGTTGTCTCAAAACCTCCTTGTAACATCTTATATGATTTATCTAATGAACTGGTGTGACTTCAAGAAATTCCCCTTTTTAATCGCCGAACAGGATGAAACTGTGATAAGTCTACGAGAGGATATAATAACATCCAAACATCTCTGTGTTATAATTGACTTATACGCAAATCACCATAAACCTCCTTGGATAATAGATCTAAACCCACAAGAAAAAACATGTGTACTGCATGACTTTATTTCTAAATCTAGGCAGGTGGACACGTCCTCCAGATCATGGAATACTTCTGACCTGGATTTGGTAATATTCTATGCATCTTTGACTTATTTGAGAAGAGGTATAATAAAACAATTAAGGATAAGACAAGTTACTGAGGTTATAGATACCACATCAATGTTAAGGGATAATATAATTGTAGAAAATCCTCCTAGTAAAACAGGAGTGTTAGACATCAGAGGTTGTATAATATACAATTTAGAGGAAATCCTGTCTATTAACACAAAATCAGCGTCAAAAAAGATCTTTAATCTTAATAGTAGGCCGTCAGTGGAGAATCATAAATATAGAAGGATAGGTCTCAACTCAACATCGTGTTCCAAGGCATTAAATCTATCACCTCTAATTCAAAGGTATCTGCCGTCAGGAGCTCAAAGGTTGTTTATAGGAGCAGGTTCTGGGAGAATGCTGTTATTATATCAGTCTACATTGGGGCAATCAATTTCTTTTTACAATTCAGGTATAGATGGAGATTATATACCAGGTCAAAGAGAACTGAAACTATTTCCCTCTGAATACTCAATTGCTGTGGAAGACCCATCTCTGACGGGGAAATTGAAAGGACTAGTGGTGCTCTTATTCAATGGGATACCAGAAACAACATGGATCGGGGATATAGTCTCCTGCGAGTATATCTGAAATAGGACAGCGAGGCGAAGTATAGGTCTTGTCCATTCTGACATGGAGTCTGGGATTGACAAAAATGTAGAGGAGATACTAGTAGGACATTCCCATCTAATATCTATGGCGATAAATGTTATGATGGAGGACGGACTATTAGTATCCAAGAGAGCATACACCCATGGATTCCCAATCTCAAGATTATTTAACATGTACAGATCATATTTTGGACTAGTACTGGTGTGTTTCCCAGTGTATAGTAATCCAGATTCTACTGAGGTATATCTTCTTTGCTTAGAAAAGACGGTCAAGACTATTGTTCCCCCGCAAAAAGTCCTTGAGCACTCTAATTTGCACGATGAAGTCAATGACCAGGGGATAACATCAGTGATTTTTAAAATCAAGAATTCACAGTCTAAGCAGTTCCACGATGATCTAAAGAAGTACTATCAGATTGACCAACCTTTTTTTGTACCAACTAAATTCACTAGTGATGAACAAGTACTTGTCCAAGCAGGGCTGAAACTCAATGGGCCAGAAATTCTTAAGAGTGAAATCAGTTATGATATCGGTTCAGATATCAATACATTAAGAGACACCATCATAATTATGTTAAATGAGGCTATGAATTATTTTGATGACAACAGATCACCTTCACACCACCTAGAACCCTATCCAGTTTTGGAGAGAACTAGAATTAAAACAATAATGAATCGTGTGACTAAGCAAGTGATTGTCTACTCACTTATCAAGTTCAAGGCCACCAAAAGTTCAGAACTCTACCACATTAAAAATAACATCAGAAGAAAAGTTCTAATCTTAGATTTCAGATCAAAGCTCATGACAAAGACTCTACCTAAAGCAATGCAAGAGAGAAGAGAAAAAAGCGGTTTCTAAGAGGTTTGGATAGTAGATTTATCGAATCGAGAAGTTAAAATCTGGTGGAAGATATTCGGATACATATCCCTTATCTGATTTAACCTTCCAAATCCAAGTCCCACTGATAACTTATGTTGATCTAAGGTTCAGTTATTAAGAAAAACTTAATAACGATTCTTCGTTACCCTTG
Navigate in a web browser to NCBI BLAST. You should see the following screen:
Click on the button for “Nucleotide BLAST”. This will redirect you to the below webpage.
To input the query, copy and paste the contig sequence into the box (this can include the header, but does not need to).
Default settings should be fine in this first instance (this will search all non-redundant records on NCBI). Click BLAST to run the search.
You may need to wait a few minutes, but results should show up automatically.
Questions
1. What are the top BLAST hits for your query sequences?
2. How confident are those hits?
3. How similar are they to something in the NCBI database?
Characterising and contextualising the initial cases
outbreak.seq_run_1.fasta file?
> characterGiven the BLAST hits, it seems likely that the sequences are Henipavirus nipahense (NiV) genomes. To confirm this, and to investigate how the samples fit into the context of known NiV diversity, we will acquire a background dataset. For this we will use the NCBI Virus resource, which is a community portal for viral data that is archived on Genbank.
Navigate to NCBI Virus and click on Search by virus name.
You can begin typing in Nipah and the species name should appear.
Click on the species name (Henipavirus nipahense) and this will redirect you to the NiV records.
Questions
1. How many records are there?
2. Would we want to download all of them? (Hint: look at the sequence lengths)
These records can be filtered by length, completeness and a variety of other factors as shown below.
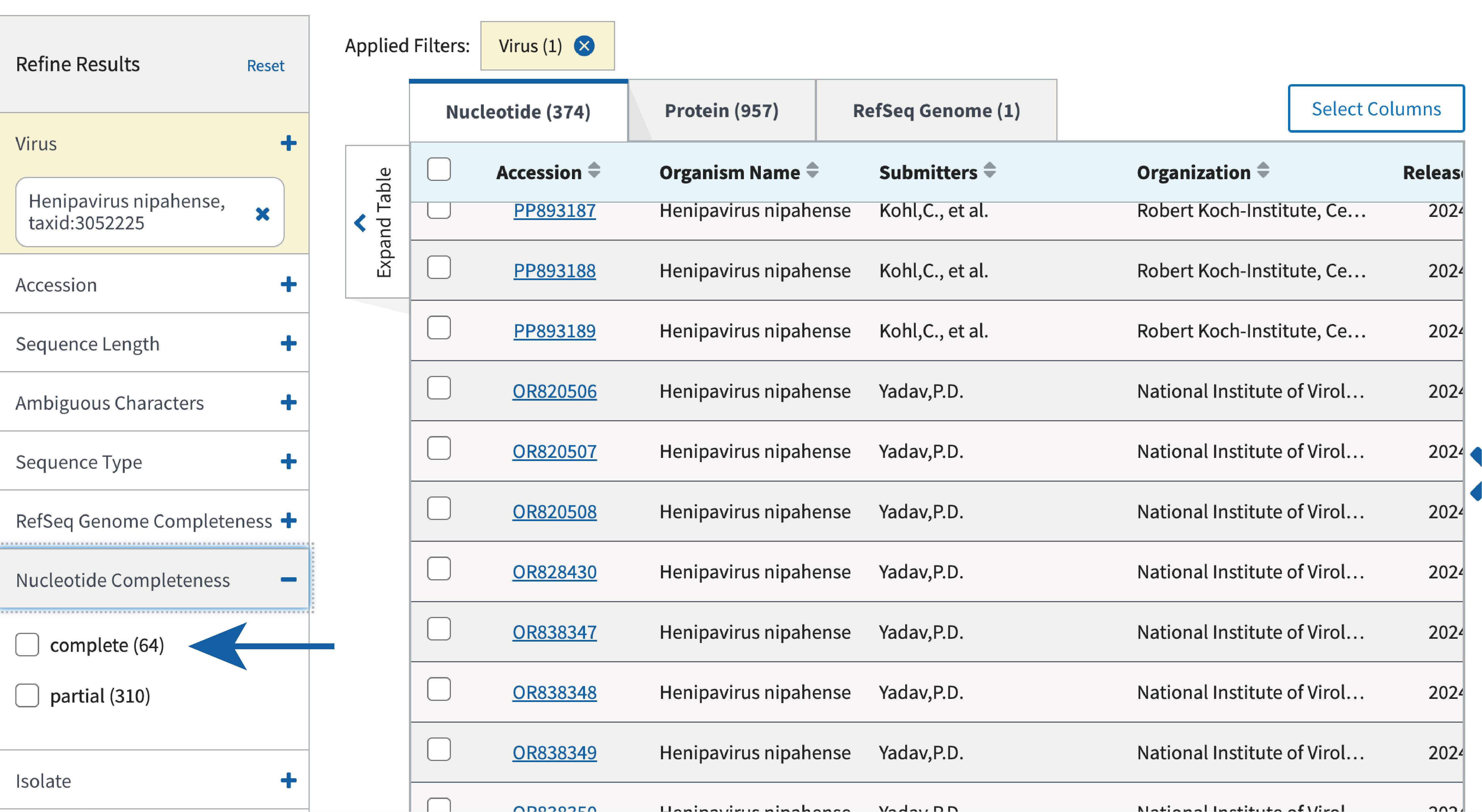
For the purposes of this exercise, we have already prepared all the complete records available for NiV that had a date and location of collection. This background dataset has been curated and headers annotated with consistent fields:
>Accession|Virus|Country|Host|CollectionDateFor example:
>MH523641|NiV|India|human|2018-05-21
This file has both the initial case sequences and the background dataset combined into one file. If actually creating this dataset, you would need to combine these files yourself (either copy & paste, or through the command line).
However, ordinarily you can download the dataset by clicking the Download button indicated below:
The fields can be customised when Downloading from NCBI Virus, however it is always a good idea to curate the headers of your dataset.
- Best practice for phylogenetics is to delimit the FASTA header fields with
|- Ensure date formats are in ISO YYYY-MM-DD format, and best practice is to have them as the last field in the header
- Do not put whitespace (spaces) into a FASTA ID. It is possible to annotate a FASTA file with a description, however this may be lost during phylogenetic analysis.
- Avoid overly long FASTA sequence headers as they make it difficult to read a phylogeny. To shorten headers, remove redundant information and consider possible contractions (e.g. NiV instead of Henipavirus nipahense).
- Avoid special characters in a FASTA sequence header. This includes but is not limited to:
,, ;, (, ), [,], &. The reason for this is that many of these are characters used to write out a phylogenetic tree string and although many software will handle this, it may be a source of unexpected error!Aligning the sequences
We now have our combined dataset, however before any trees can be built, we need to align them. Feel free to align with your preferred method, however this tutorial will use the MAFFT online version for alignment. MAFFT has a variety of different algorithms, with varying speeds and precision. Today we’ll just be using the AUTO option that selects the most appropriate for your input dataset. For full details about MAFFT and its models, see Katoh et al 2002.
Navigate to the MAFFT web server.
We have provided the combined case genome sequences and NiV background set. Once you have decompressed the outbreak.seq_run_1.NiV_background.fasta file, select Choose file on the MAFFT web server and upload your file. We will run with the default AUTO mode (it may be useful to change the Title length field to 50, but not necessary), select Submit.
When ready, return to the results page and click on Fasta format to download the alignment as a FASTA file.
The downloaded file will probably have a long and uninformative name such as: _out.2502241929466CMs0BqwdZEKASUk69JNo3lsfnormal.fasta – rename this file to outbreak.seq_run_1_aligned.fasta
Estimate a Maximum Likelihood tree
For the initial exploratory analysis, we want to quickly estimate a phylogeny to place our sequences into the known diversity of NiV. In order to do this, we will use IQTREE to estimate the phylogeny Minh et al. 2020. IQTREE2 can be run on the command line with more flexibility, but we will be using the web server in this tutorial Trifinopoulos et al. 2016.
To navigate to the web server, click the W-IQTREE link.
Input data
Upload your alignment file to the server by clicking Browse and selecting the file outbreak.seq_run_1_aligned.fasta. You can select DNA as the sequence type but.
Substitution model options
We will bypass model selection for now, and just select the simple HKY substitution model. This is just a quick first look at the data.
Running IQTREE
All other parameters can be left as default. Scroll down and select Submit job.
Click QUERY STATUS to check on the run. When the job has finished the status bar on the left will say Success.
You can examine an ASCII version of the phylogeny in the Full Result tab, but to download the result, click DOWNLOAD SELECTED JOBS in the bottom left. Decompress the downloaded file and you’ll see the contents contain the IQTREE log file, the result file and the treefile.
We will use FigTree to look at the treefile (the file ending in .treefile).
Open the FigTree application and select to open the treefile, it will be displayed as arbitrarily rooted because we did not specify an outgroup during the quick run. Click on the Tree dropdown menu on the top bar and select Midpoint Root.
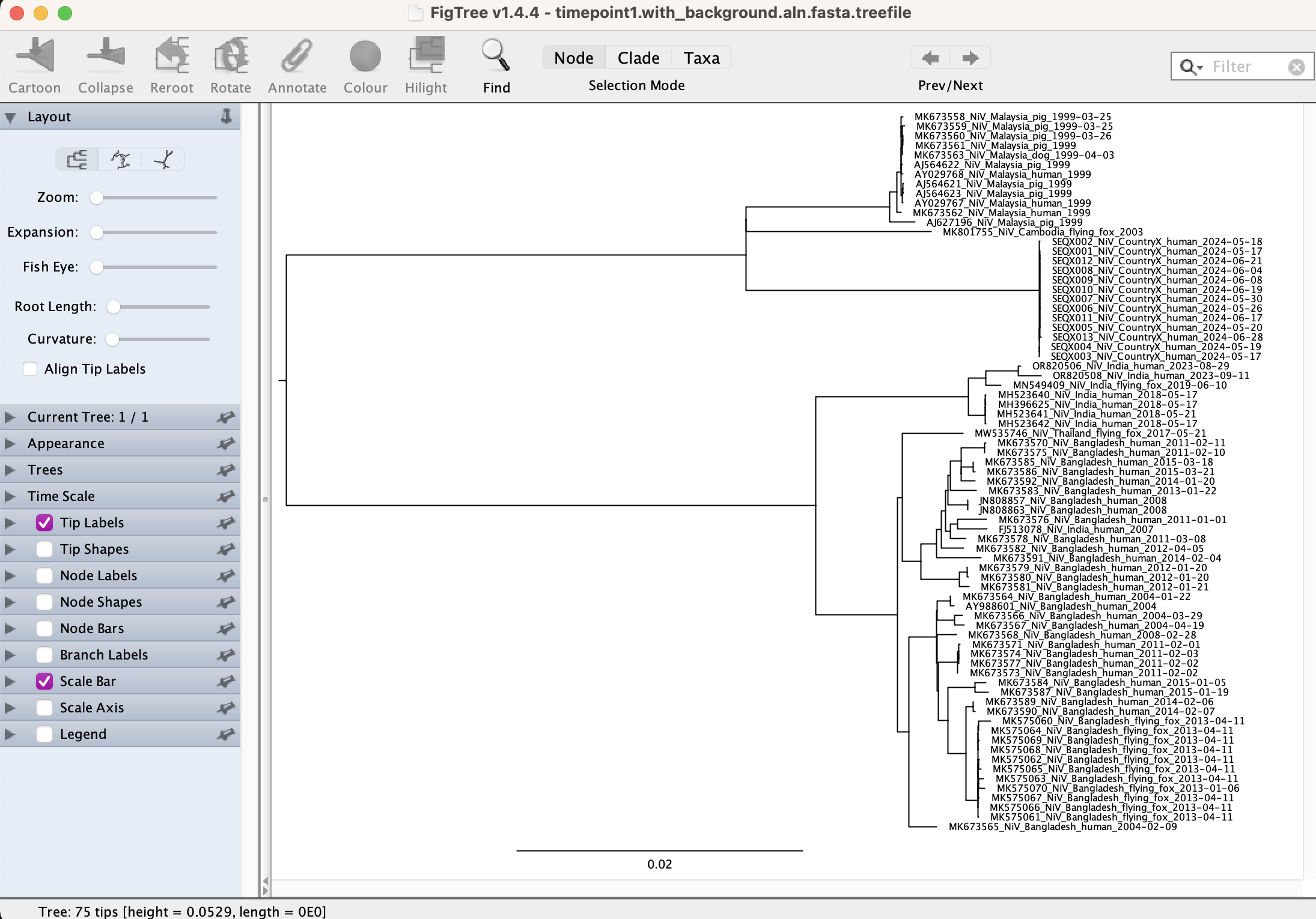
This will re-root the phylogeny and provide a more balanced tree view in the absence of a known outgroup.
|) to underscores (_). This is fine for our exploratory purposes, but know that command-line IQTREE2 does not do this and be aware that the tip-labels in the tree will now be not exactly identical to the FASTA sequence headers.Examine the phylogeny.
1. What does this phylogeny tell you about the cases? Do they cluster together?
2. Is there good support for this?
3. What can you say about the outbreak?
When did this outbreak arise?
Even with just these few sequences it may be possible to ascertain some key information about this outbreak (such as when it likely began and how quickly it is spreading).
Use the TempEst tutorial as a guide to load the tree file into TempEst to assess whether there is temporal signal in the data. Parse the tip dates by selecting Parse dates in the Sample Dates tab, and select Best fitting root in the top-left side of the application.
Take a look at the root-to-tip and residual tabs.
1. What does TempEst estimate the rate to be? Does this seem sensible?
2. What is the estimated tMRCA?
3. Is there good temporal signal?
Next, use BEAUTi to generate an XML file for BEAST to run. full step-by-step details can be found at
in the Rates and dates tutorial. Add the outbreak.seq_run_1.fasta as a partition, in the Tips tab parse the dates from the tips and set an exponential growth model. It’s a small dataset, with very little temporal signal, so on the MCMC tab set the chain length to a little longer (perhaps 100,000,000 states) and reduce logging frequency to every 10000 states.
When ready, click Generate BEAST File....
Run BEAST with your newly generated XML (detailed instructions on how to run BEAST can be found in the First tutorial).
Open Tracer and load the newly generated log file to assess the BEAST run (the Rates and dates tutorial also includes some useful tips about importing into Tracer and interpreting the plots.).
Setup... or Setup Chart. The number of bins for the histogram can also be toggled.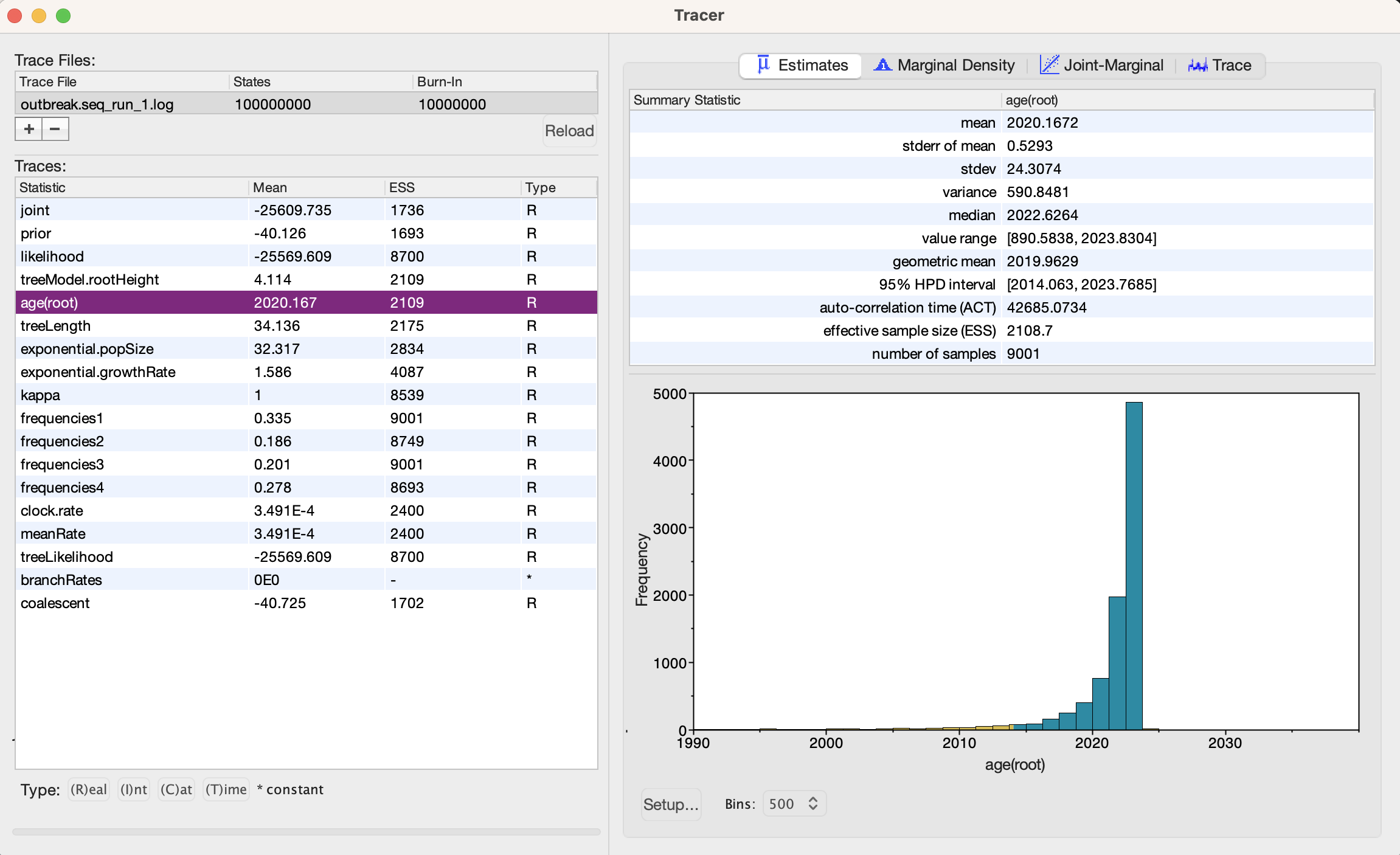
1. What is your assessment of the BEAST run?
2. What is the root age estimate? How confident is this estimate?
3. What is the clock rate estimate (and 95 HPD interval)? Does this make sense for NiV? 4. What would you suggest is needed for a better estimate?
Use Tree Annotator to produce a maximum clade credibility (MCC) tree from the .trees file and view in FigTree. Using the FigTree tutorial, set the Time Scale and display the 95 HPD for the height estimates on the nodes as shown below.
1. From the information available, what can you say about the origin of this outbreak?
EXERCISE 2: Additional human cases spurs an investigation in animal population
Download the provided FASTA alignment file and decompress. This FASTA file contains NiV genome sequences from both the initial sequencing run and a second sequencing run that contained both human and pig NiV samples. We will attempt to answer whether the human and animal outbreaks are linked and what is the source of these cases.
Generate a maximum likelihood tree from the alignment
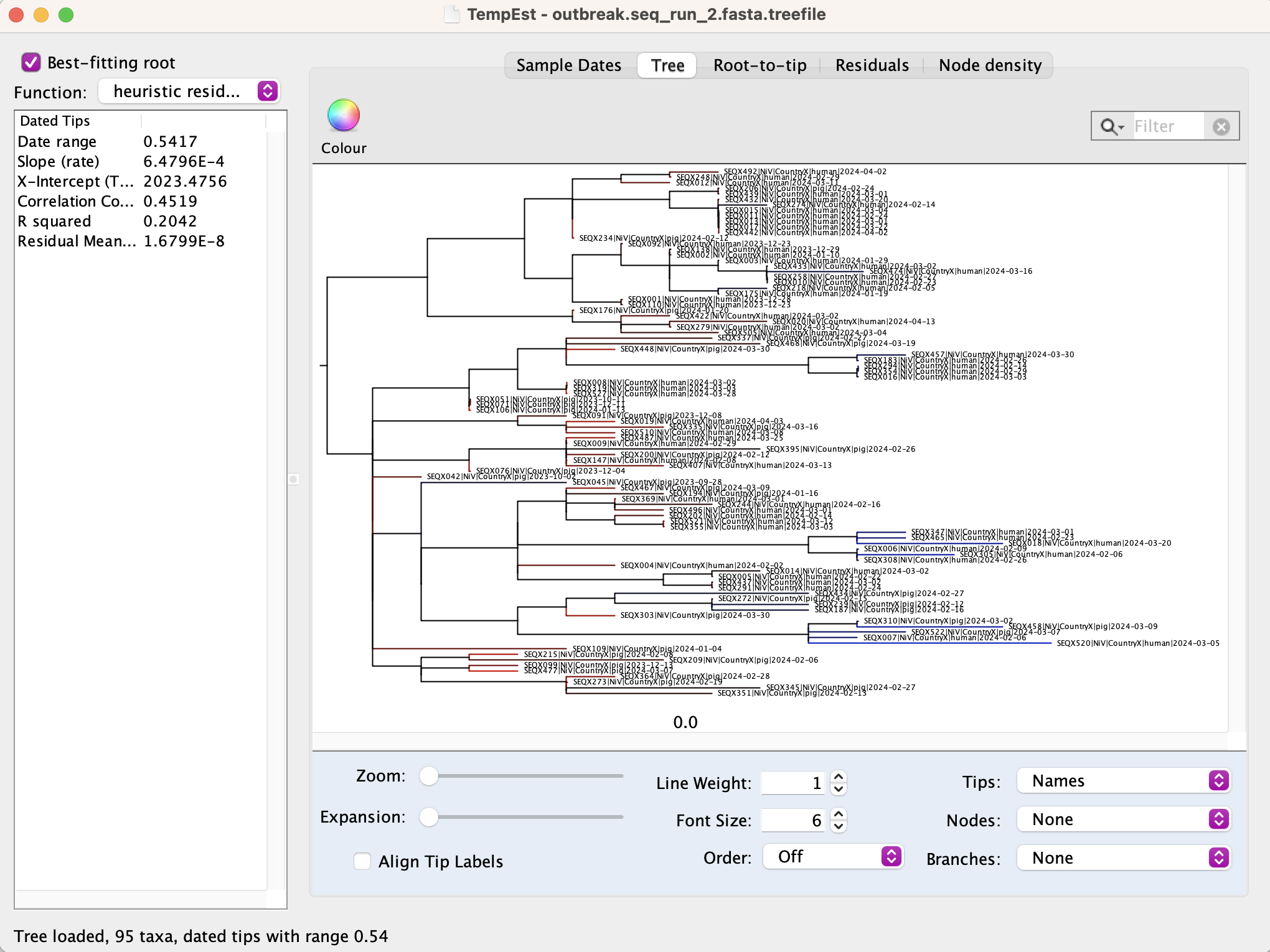
Using IQTREE as described above in EXERCISE 1, estimate a maximum likelihood phylogeny with the outbreak.seq_run2.fasta file. Inspect the phylogeny using TempEst (parse tip dates and select best fitting root). Note that the tips are labelled with host species.

1. What does this phylogeny tell us about the human and pig outbreaks?
Examine the root-to-tip and residual plots.
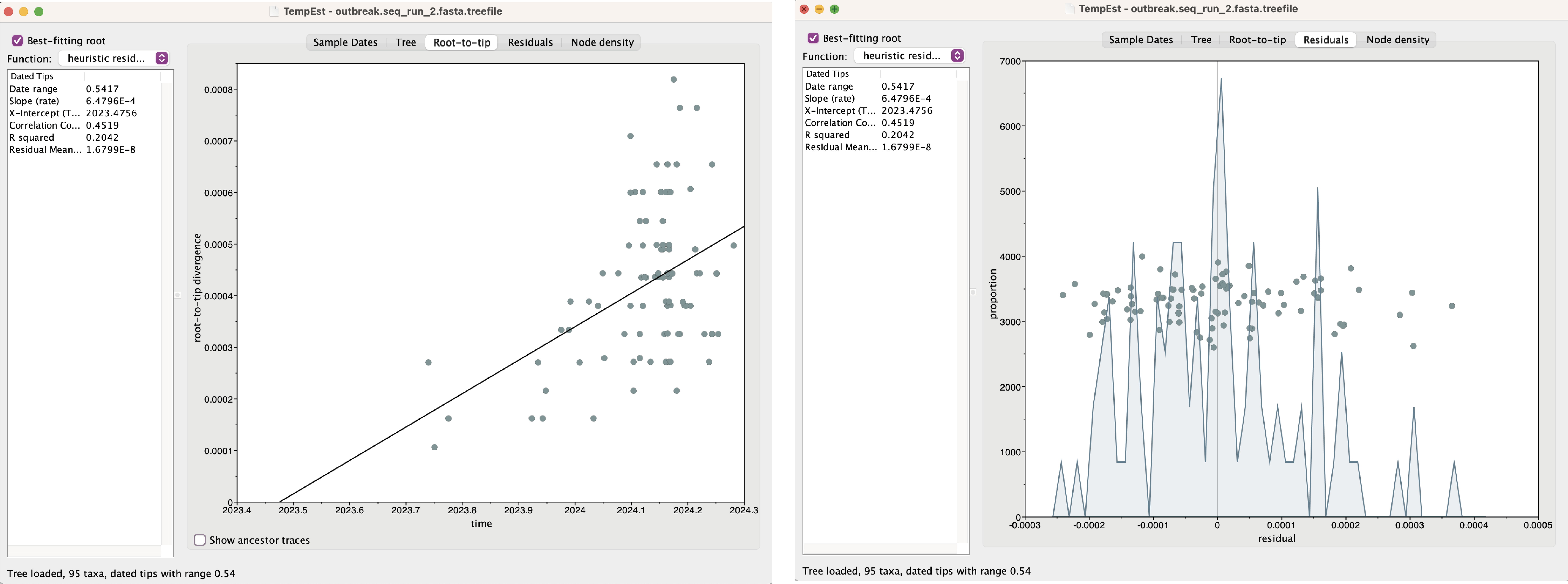
1. What is the estimated rate?
2. What is the estimated tMRCA?
Estimate a time tree and reconstruct ancestral host states
Launch BEAUTi and import the outbreak.seq_run2.fasta alignment.
Parse the tip dates on the Tips tab.
Navigate to the Traits tab and create a new trait called host.
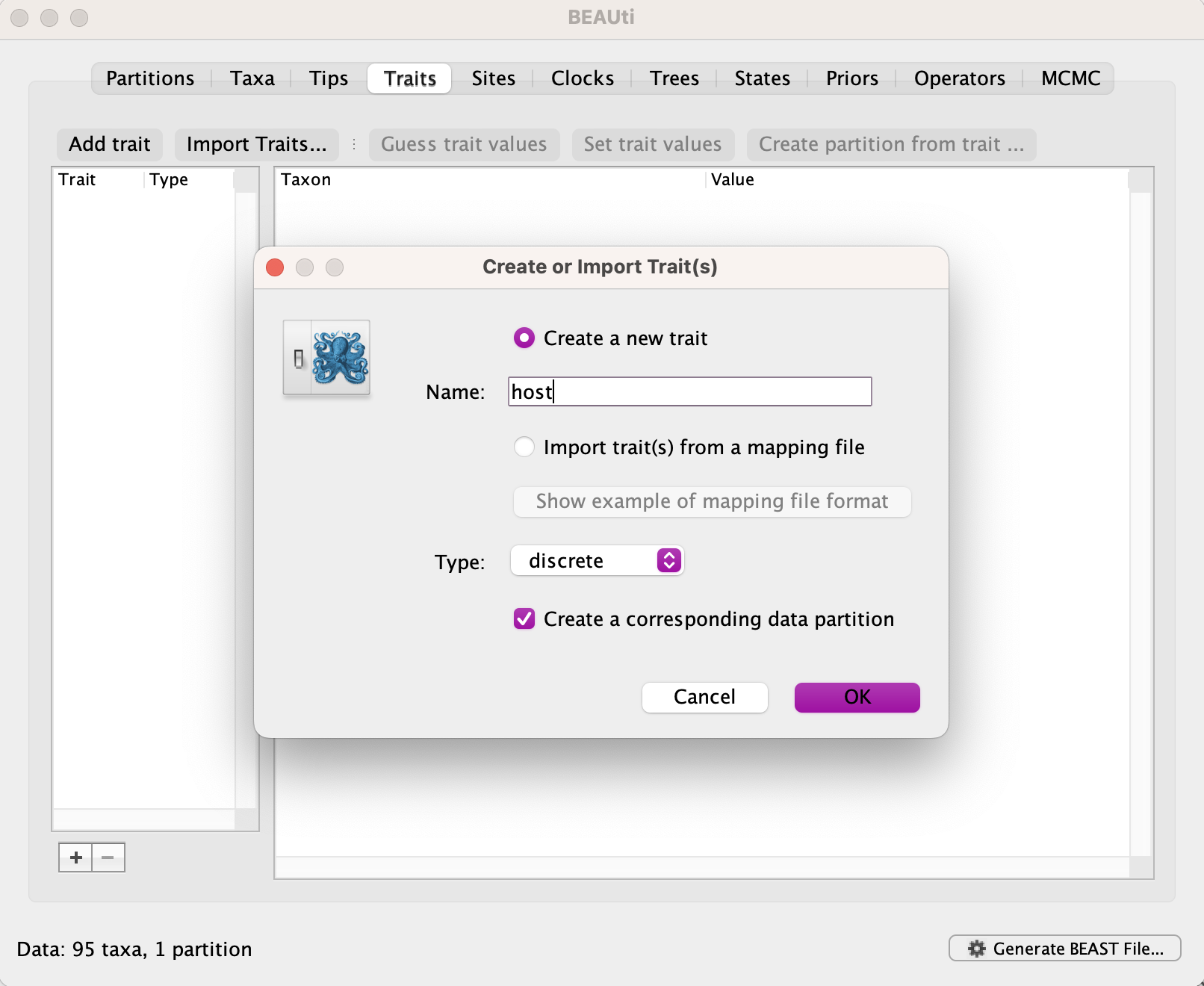
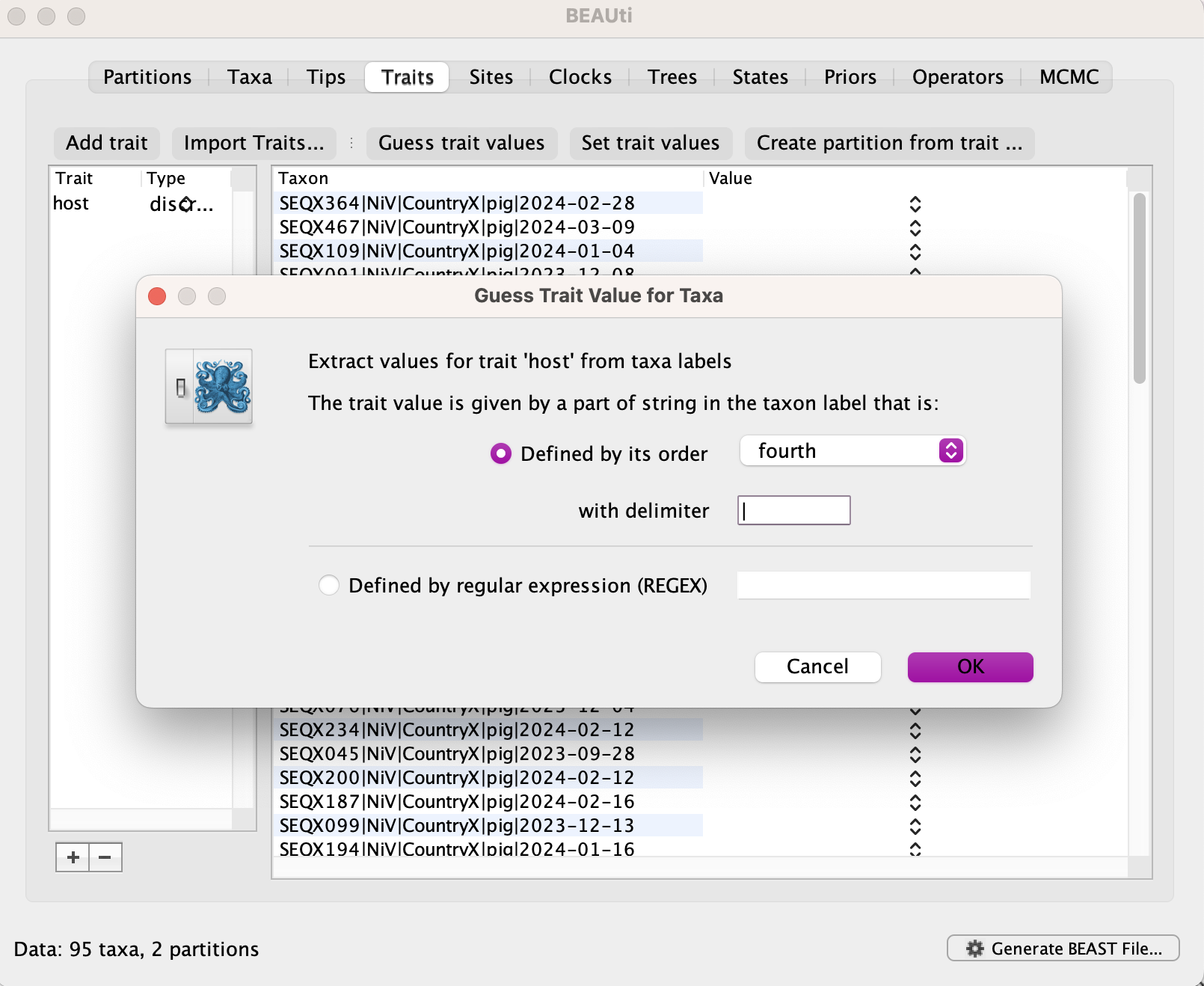
Select Guess trait values and indicate it is the fourth field in the pipe(|)-delimited header.
Create a new partition with that trait called host.
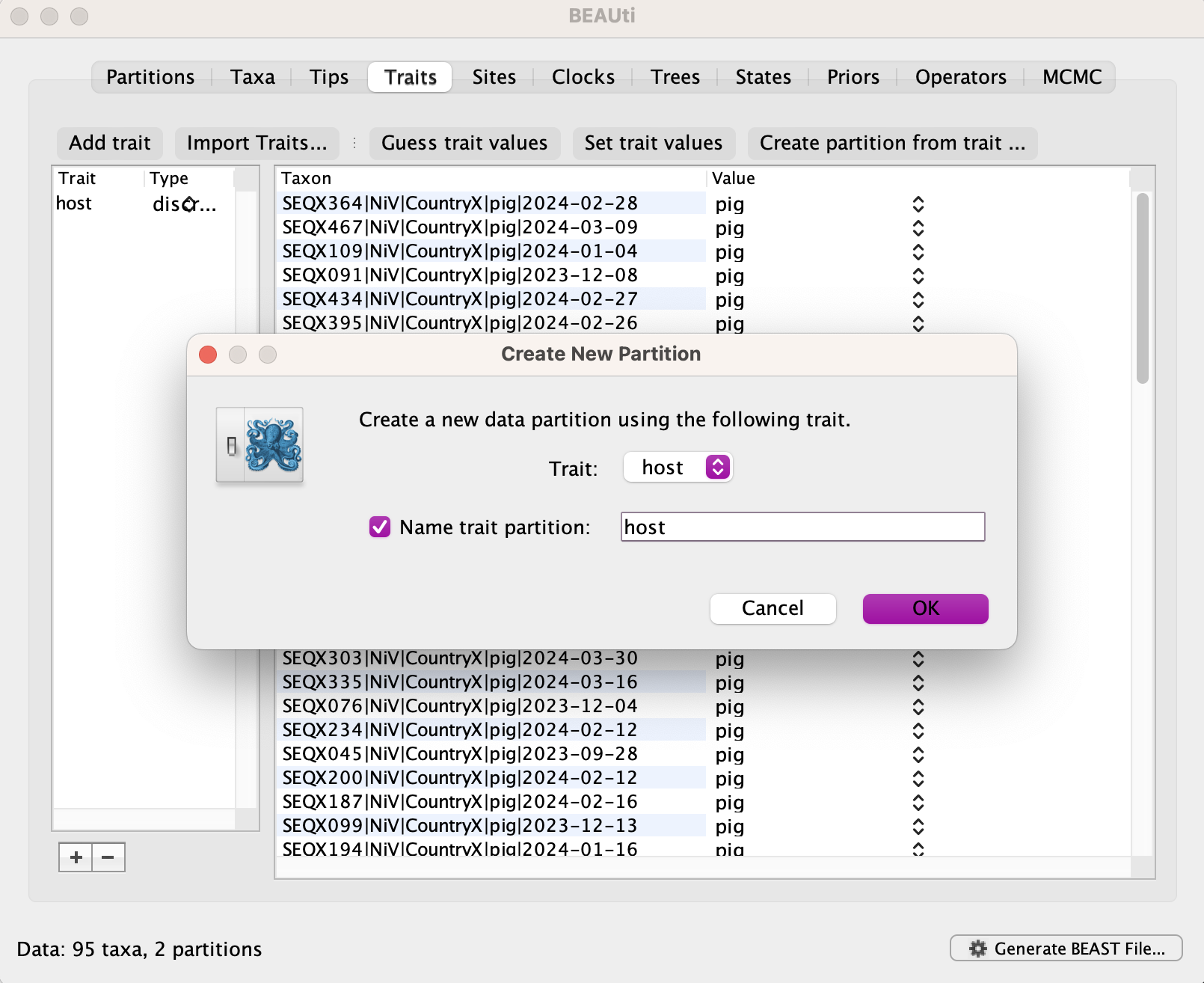
If you navigate back to the partitions tab, you’ll see host has now appeared beneath the nucleotide partition.
Click on host in the Substitution model window (Sites tab) and keep the Discrete Trait Substitution Model as Symmetric substitution model but select the option to perform BSSVS (Infer social network with BSSVS). The symmetric substitution model specifies a discrete state ancestral reconstruction using a standard continuous-time Markov chain (CTMC), in which the transition rates between locations are reversible. Selecting the BSSVS option enables the Bayesian Stochastic Search Variable Selection procedure. This procedure will attempt to limit the number of rates (at least k-1, where k is the number of states) to only those that adequately explain the phylogenetic diffusion process.
In the Trees tab, we will set the Tree Prior to an Exponential Growth coalescent model.
Ensure under the States tab that the host partition is set to reconstruct the states of all ancestors (should be on by default).
We are now ready to create the BEAST XML file. Select Generate XML....
Run BEAST using the newly generated XML as before.
Examine the output logfile in Tracer.
1. Has the increased sampling improved the BEAST run output?
2. What is the estimated tMRCA? Look at the 95 HPD, has the confidence in the estimate improved with additional data?
Combine the logged trees into a MCC tree using TreeAnnotator, check if the default burn in of 1,000,000 states is sufficient in Tracer. Vizualise the MCC tree in FigTree. Set the time scale on the X axis and toggle on the Node bars showing height 95 HPD.
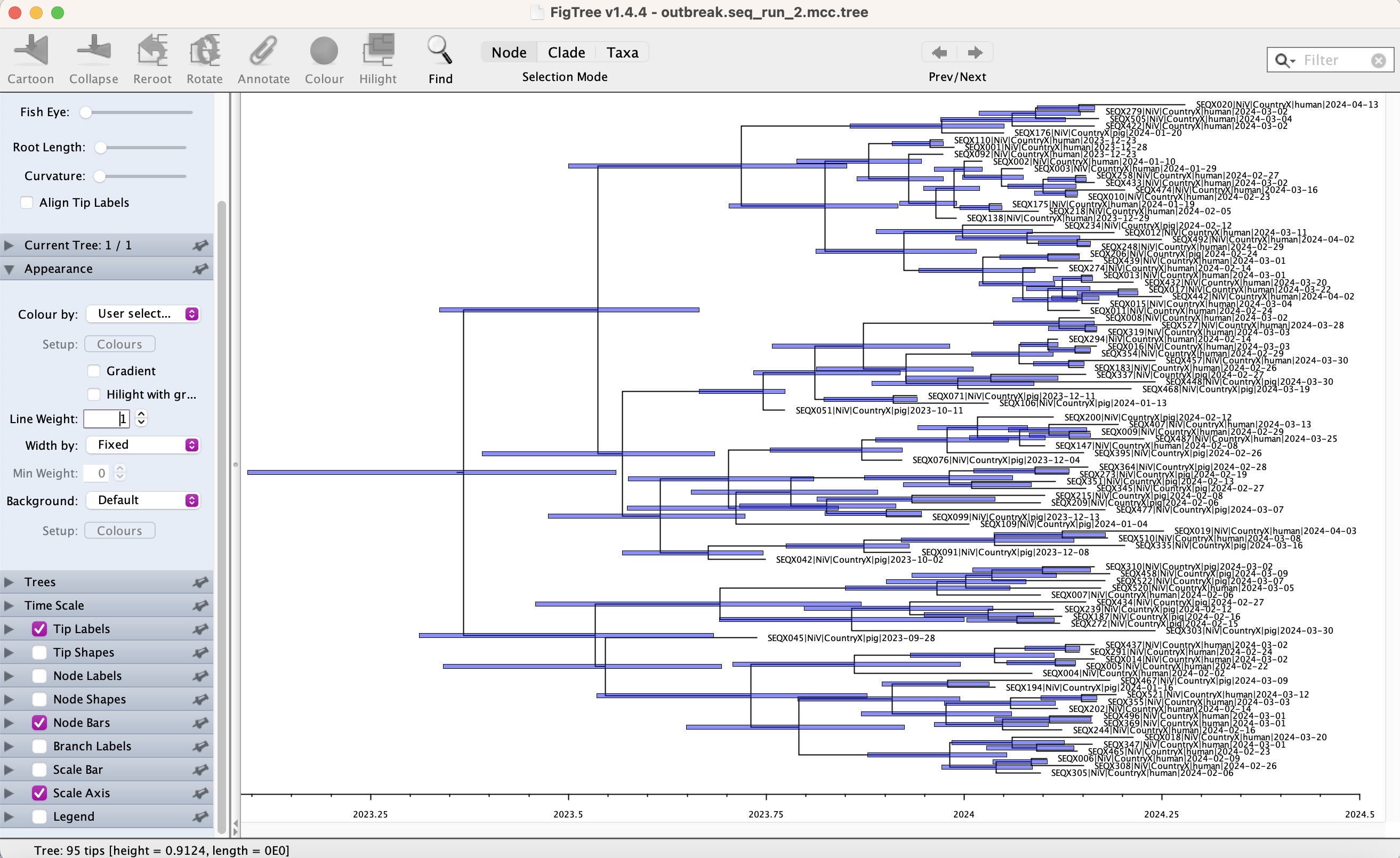

1. Has the root age HPD improved? Does it agree with the
TempEst estimation?Colour the branches of the time tree by host. Look at the confidence values for that reconstruction.

1. What does the reconstruction tell us?
2. Does it make sense?
3. What factors may have impacted this reconstruction?
Simulation Notes
An outbreak structure was simulated using JT McCrone’s transmission simulator. The simulator generated an outbreak with a total of 517 cases and produces a transmission tree, a time tree and a line list with sampling time in days.
As an example case, NiV was selected because it is fast evolving and has a suitable background dataset available on Genbank that is not too large that it would require downsampling. All complete NiV genomes were downloaded from NCBI virus, aligned using MAFFT and an ML tree was computed using IQTREE2.
Ancestral reconstruction was carried out also using IQTREE and the genome sequence for the common ancestor node for MK801755 and the 1999 porcine outbreak in Malaysia. From this ancestral node, seq-gen was used to simulate a branch length of 0.018 under the HKY substitution model, which represents ~40 years of NiV evolution assuming a rate of 4.5x10^-4 substitutions per site per year (as published for NiV in Cortes-Azuero et al 2023). This provided the initial infection case from which we simulated genome sequences corresponding to the simulated outbreak time tree. The evolution was simulated with a HKY substitution model, also using seq-gen, at a rate of 7x10^-4. The time tree was manually labelled in FigTree with host annotations, creating a scenario where pig represented the majority of cases, including the earlier cases, and human clusters represented spillover events from the pig population. In total, 60 of the 513 tips were labelled as human cases and the remainder were labelled as pig. The start date of the outbreak was set to 2023-08-11.
Because this outbreak has been simulated, we know the underlying ground truth and can see the impact lack of data and sampling bias can have on inference. Below is the BEAST DTA inference with all of the simulated outbreak cases (so 100% of the population has been sequenced and sampled in this case).
References & Further Reading
- Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214.
- Drummond AJ, Ho SYW, Phillips MJ & Rambaut A (2006) PLoS Biology 4, e88.
- Drummond AJ, Rambaut A & Shapiro B and Pybus OG (2005) Mol Biol Evol 22, 1185-1192.
- Drummond AJ, Nicholls GK, Rodrigo AG & Solomon W (2002) Genetics 161, 1307-1320.
- Dudas G, Carvalho LM, Rambaut A, Bedford T (2018) MERS-CoV spillover at the camel-human interface eLife 7:e31257.
- Ferreira, M. A. R. and M. A. Suchard. 2008. Bayesian analysis of elapsed times in continuous-time Markov chains. Can J Statistics, 36: 355–368. doi: 10.1002/cjs.5550360302
- Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, and Suchard MA (2013) Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol 30, 713-724.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013 Apr;30(4):772-80. doi: 10.1093/molbev/mst010. Epub 2013 Jan 16. PMID: 23329690; PMCID: PMC3603318.
- Minin VN, Bloomquist EW and Suchard MA (2008) Smooth Skyride through a Rough Skyline: Bayesian Coalescent-Based Inference of Population Dynamics. Molecular Biology and Evolution 25:1459-1471; doi:10.1093/molbev/msn090.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015 Jan;32(1):268-74. doi: 10.1093/molbev/msu300. Epub 2014 Nov 3. PMID: 25371430; PMCID: PMC4271533.
- Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC (2008) The genomic and epidemiological dynamics of human influenza A virus. Nature, 453: 615-9.
- Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JSM, Guan Y & Rambaut A (2009) Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122-1125.
- Trifinopoulos J, Nguyen L, von Haeseler A, Minh B, W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis, Nucleic Acids Research, Volume 44, Issue W1, 8 July 2016, Pages W232–W235, https://doi.org/10.1093/nar/gkw256
Help and documentation
The BEAST website: http://beast.community
Tutorials: http://beast.community/tutorials
Frequently asked questions: http://beast.community/faq