User-interface
Double-clicking the TreeAnnotator icon will result in a dialog box appearing:
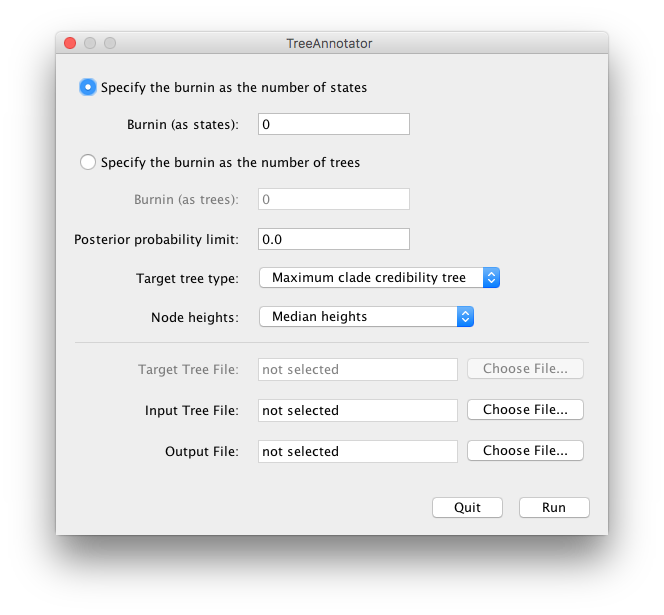
The options
BurnIn
This option allows you to select the amount of burn-in, i.e., the number of samples that will be discarded at the start of the run, so that you are only analysing the part of the trace that is in equilibrium. If the file has been processed through LogCombiner then the burnin may have already been removed. There are two ways of specifying this:
- BurnIn (as states)
- With this option you specify the number of states in the MCMC chain you wish to discard as a burnin. This will be irrespective of the sampling frequency.
- BurnIn (as trees)
- In this case you just specify the actual number of trees you wish to discard rather than the number of states.
Posterior probability limit
Posterior summaries will only be calculated for the nodes in the target tree that have a posterior probability greater than the specified limit. This is a way of removing summary statistics from nodes with very little support where these may be unreliable.
Target tree type
If you select the Maximum clade credibility option then the node height and rate statistics will be summarized on the tree in the posterior sample that has the maximum sum of posterior probabilities on its n − 2 internal nodes.
See this page for a description of the MCC trees and various other summary trees.
If you select the “User target tree” then the tree statistics will be summarized on a user-specified tree.
Node heights
This option allows you select how the node heights are summarised on the target tree. You can choose to keep the heights that the target tree has, or rescale it to reflect the posterior mean/median node heights for the clades contained in the target tree.
Target tree file
This option allows you to select and input the target tree.
This option will only be available if you have selected User target tree from the Target tree type combo menu.
Input tree file
This option allows you to select the input tree file (the tree file produced by a BEAST analysis).
Output file
This option allows you to specify a new file that the summarized tree file will be saved to.
Once ready to run the dialog box will look something like this:
Running TreeAnnotator
When you click Run, TreeAnnotator will start to summarize the tree data produced by BEAST:
TreeAnnotator v1.10, 2002-2017
MCMC Output analysis
by
Andrew Rambaut and Alexei J. Drummond
Institute of Evolutionary Biology
University of Edinburgh
a.rambaut@ed.ac.uk
Department of Computer Science
University of Auckland
alexei@cs.auckland.ac.nz
Reading trees (bar assumes 10,000 trees)...
0 25 50 75 100
|--------------|--------------|--------------|--------------|
************************************************************
Total trees read: 10000
Ignoring first 1000000 states (1000 trees).
Total unique clades: 85
Finding maximum credibility tree...
Analyzing 9000 trees...
0 25 50 75 100
|--------------|--------------|--------------|--------------|
************************************************************
Best tree: STATE_1091000 (tree number 1092)
Highest Log Clade Credibility: -5.1142294006748275
Collecting node information...
0 25 50 75 100
|--------------|--------------|--------------|--------------|
************************************************************
Annotating target tree...
Writing annotated tree....
Finished - Quit program to exit.
TreeAnnotator will state the number of trees that have been read, will find the best fit tree specified under the “Target tree type”.
The progress of this will be monitored by the * symbols moving across the screen.
When the program says ‘Finished’, the annotated tree file will have been written. Close the program by selecting the Quit option.
The tree file can then be displayed in FigTree.
Command-line use
To run treeannotator on the command-line you can use the shell script installed in /bin of the BEAST package. Use the help command to see a list of options:
bin/treeannotator -help
TreeAnnotator v1.10, 2002-2017
MCMC Output analysis
by
Andrew Rambaut and Alexei J. Drummond
Institute of Evolutionary Biology
University of Edinburgh
a.rambaut@ed.ac.uk
Department of Computer Science
University of Auckland
alexei@cs.auckland.ac.nz
Usage: treeannotator [-heights <keep|median|mean|ca>] [-burnin <i>] [-burninTrees <i>] [-limit <r>] [-target <target_file_name>] [-help] [-forceDiscrete] [-hpd2D <the HPD interval to be used for the bivariate traits>] <input-file-name> [<output-file-name>]
-heights an option of 'keep' (default), 'median', 'mean' or 'ca'
-burnin the number of states to be considered as 'burn-in'
-burninTrees the number of trees to be considered as 'burn-in'
-limit the minimum posterior probability for a node to be annotated
-target specifies a user target tree to be annotated
-help option to print this message
-forceDiscrete forces integer traits to be treated as discrete traits.
-hpd2D specifies a (vector of comma seperated) HPD proportion(s)
Example: treeannotator test.trees out.txt
Example: treeannotator -burnin 100 -heights mean test.trees out.txt
Example: treeannotator -burnin 100 -target map.tree test.trees out.txt
Notes:
-
The last two options are specific to summarising trees with discrete and continuous trait values and will be dealt with elsewhere.
-
Just typing
treeannotatorwith no additional options will bring up the user-interface dialog box.