By having sequences sampled through time, BEAST is able to estimate the timescale of the tree and the rate of evolution. See Rambaut (2000) for a description of how this is done (albeit in a maximum likelihood rather than Bayesian context). See Dummond et al 2003 for a review of the idea of ‘measurably-evolving populations’.
Dates are generally specified as a calendar date such as 4 May 1971, an amount of time since some point in the past such as 200 days since diagnosis, or an ‘age’ such as 10,000 years before present. Which ever way they are specified, BEAST will convert these dates to a standard time axis that runs backwards from the most recent tip which becomes the zero point in the axis. If provided as calendar dates then the timescale will be in units of years. For other forms of dates the units of time will be that of the dates provided - days, years, thousands of years etc. Within BEAST, times on this timescale are referred to as ‘heights’ — i.e., root height, node heights etc. — which comes from terminology in computer science where trees are drawn with tips at the bottom of the page and the root at the top so its time scale is how high up the page it is.
Importing dates into BEAUti
When a data set has been loaded into BEAUti, the Tips panel will show a table with all the taxon labels and their assigned dates. By default all the taxa are assumed to have a date of zero. This will be correct if all the sequences were sampled at approximately the same point in time (at least on the time scale of their evolutionary history). Click the option Use tip dates to tell BEAUti that you wish to use them. At this point all the controls will become available and you can provide dates for the sequences. If you loaded this file (an alignment of 35 RSV viruses) the table would look like this:
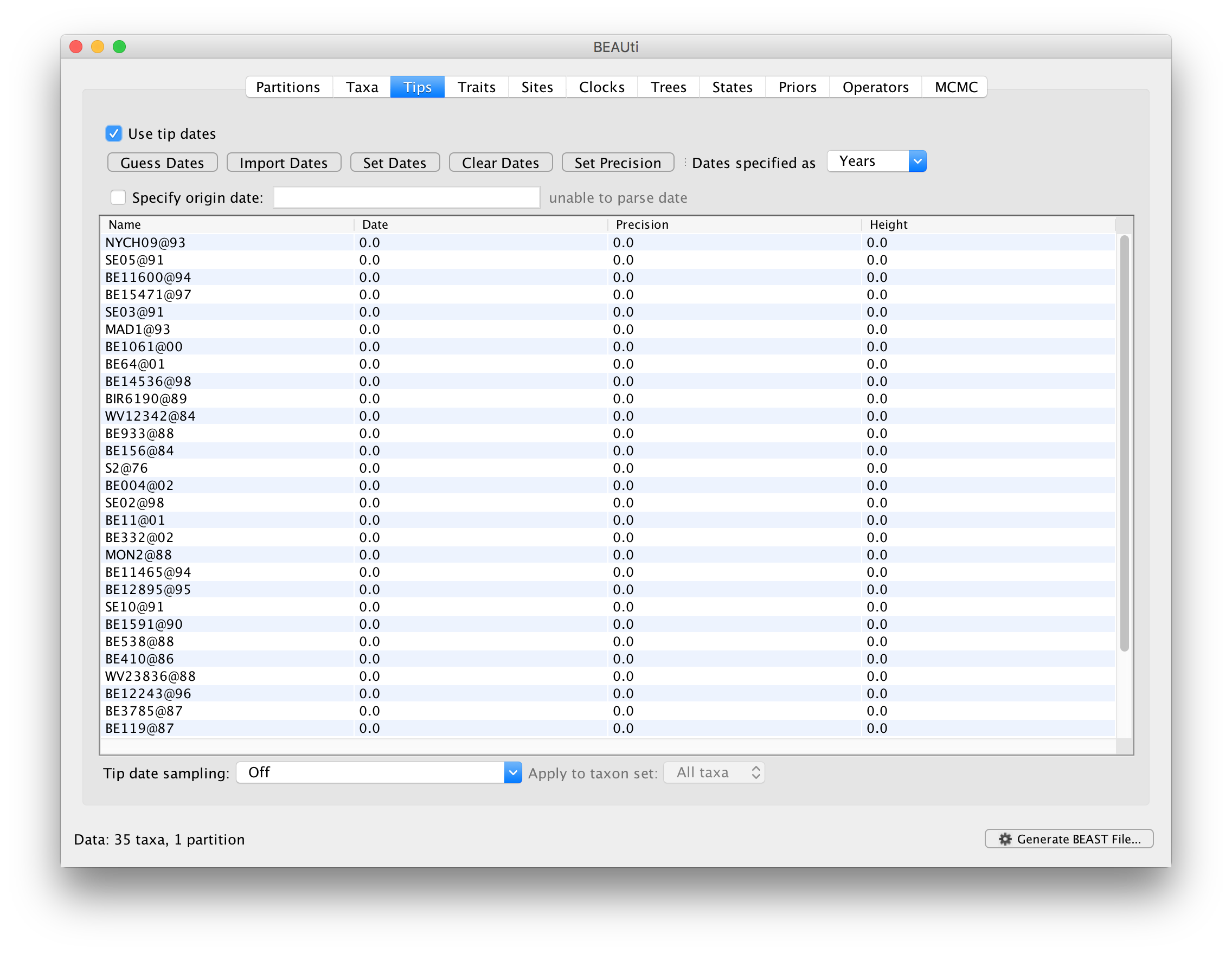
BEAUti provides a number of ways of loading dates for the sequences of an alignment and a great deal of flexibility in the format of those dates. In this case the dates are given as two digit years prefixed by an @ symbol. This is not a standard but just how the creator of the data chose to label these sequences. The most straight forward (but laborious) way of setting the dates is simply to edit the value in the table:
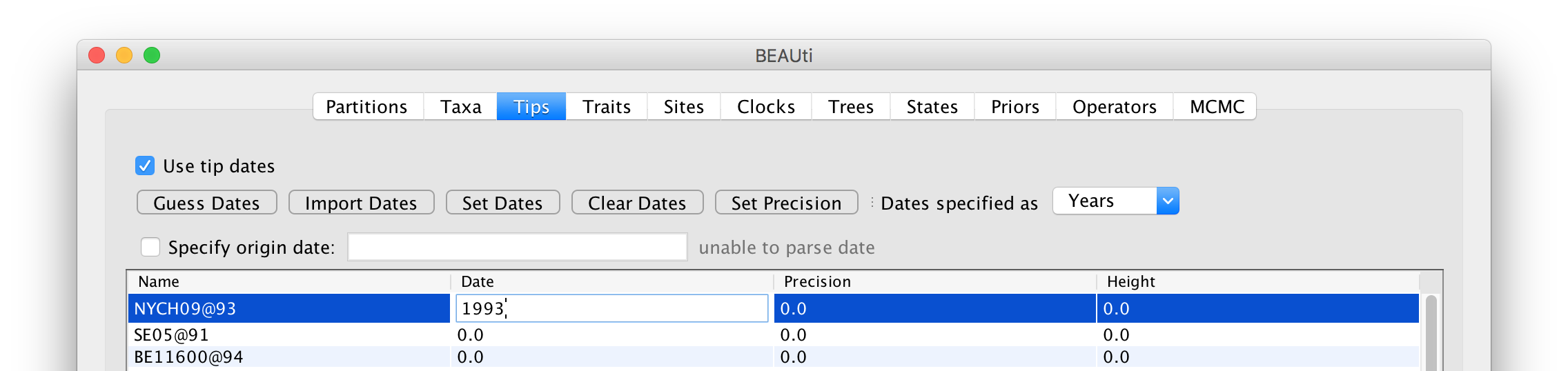
Here the two digit year was 93 referring to 1993 (presumably not 1893 or though this is not implausible) so we enter that value in the date column. In this column BEAST is expecting dates on a continuous time-line so it should be in decimal years (i.e., months are 1/12 or 0.08333 of a year and days are approximately 0.00274 of a year). Once one date has been specified, BEAST’s timescale is set:
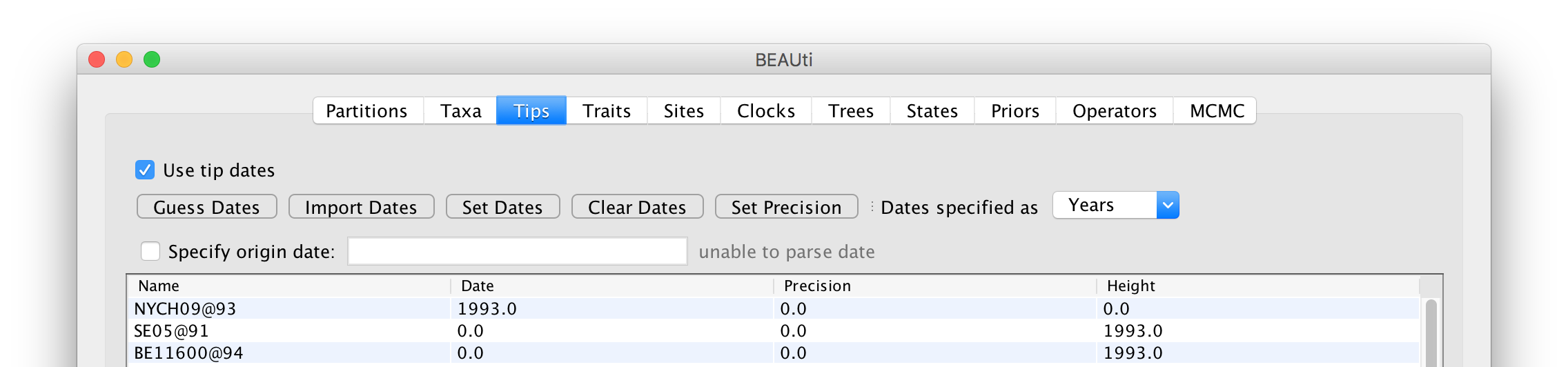
In the image above, the first sequence has been given the date 1993 and BEAST has given it the height of 0 (it is now the most recently sampled sequence). All the other sequences have a height of 1993 — i.e., BEAST thinks they have been sampled 1993 years before the first one in the year 0 (technically, in the Gregorian calendar there is no year zero but we can ignore that poor decision). So we need to set the dates for all the other sequences. For convenience when many sequences have the same date, you can select multiple rows in the table and then click the Set Dates button to set the same value to all of them. You can also Clear Dates for selected rows but that is the same as setting their date to zero.
Extracting dates from taxon labels
BEAUti can also be parsed when encoded in taxon labels (such as is the case in this RSV data set). To do this, press the Guess Dates button (probably a bit mis-named as the procedure won’t be guessing). Clicking this will make the following dialog box appear:
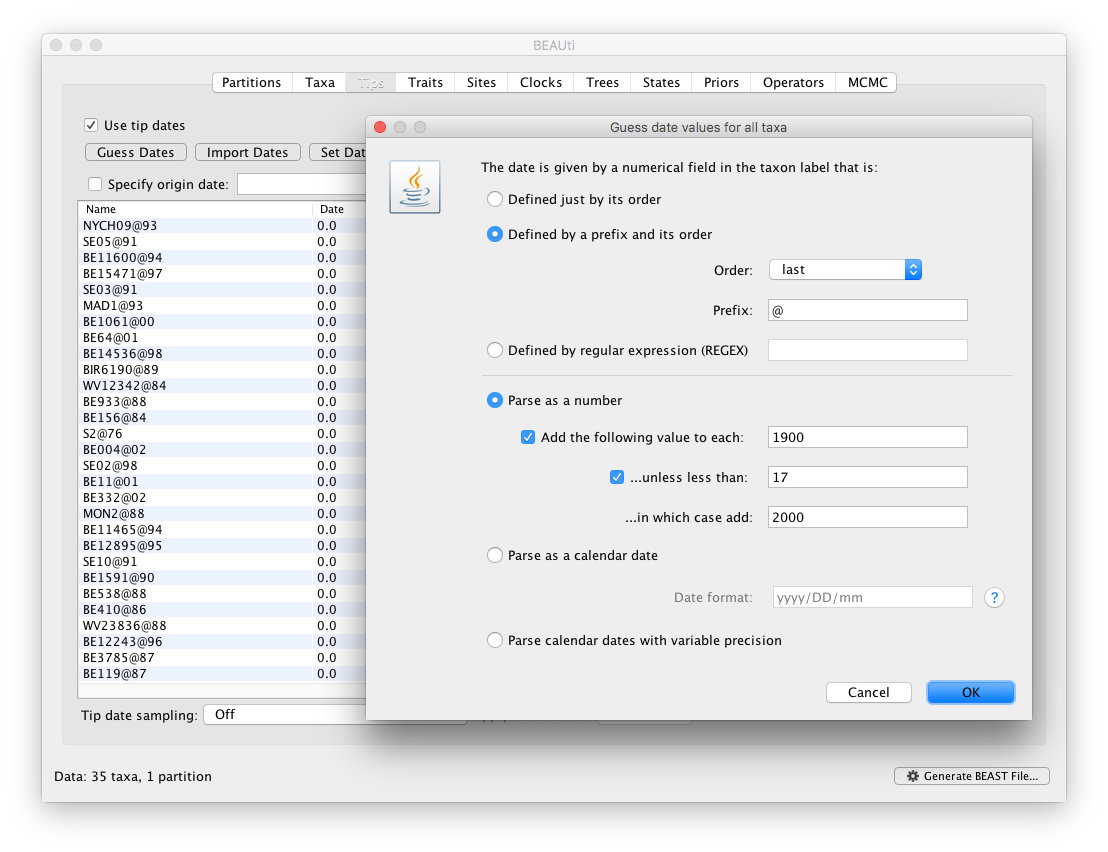
This operation attempts to guess what the dates are from information contained within the taxon names. Generally this works by trying to find a numerical field within each name. If the taxon names contain more than one numerical field (such as the RSV sequences, above) then you can specify how to find the one that corresponds to the date of sampling. You can either specify the order that the date field comes (e.g., first, last or various positions in between) or specify a prefix (some characters that come immediately before the date field in each name). For the RSV sequences you specify the prefix @.
In this dialog box, you can also get BEAUti to add a fixed value to each guessed date. In this case the value “1900” has been added to turn the dates from 2 digit years to 4 digit. Any dates in the taxon names given as “00” would thus become “1900”. If these dates were actually referring to the year 2000 then selecting the next option would convert them correctly, adding 2000 to any date less than 17.
When you press OK the dates will appear in the appropriate column of the main window. You can then check these and edit them manually as required. If you click the column heading for the ‘Date’ column it will sort the rows in ascending order of date. Click again and it will show sort in descending order. This allows you to check the extremes for mis-parsed dates.
Specifying formats for parsing dates
This dialog box also provides a wide range of other options for extracting dates from taxon labels. The first half specifies how to find the dates in the taxon labels and has 3 options.
- Defined just by its order
- This option detects entirely numerical fields (i.e. containing only digits, ‘.’ and ‘-‘) separated by non numerical characters and then uses the
Ordermenu to specify which of thes is the date field (e.g., always the first one or the second from last). - Defined by a prefix and its order
- This option finds fields that are prefixed by a particular character or string (e.g., the
@symbol, above) and then uses the order to specify which one. - Defined by regular expression (REGEX)
- A regular expression or REGEX is a flexible way of matching parts of complex strings widely used in computer science. It is beyond the scope of this document to describe the full potential of this but here is a good description. As an example, the pattern
\d\d\d\d-\d\d-\d\dwould match any date string formatted like2017-08-01(the\dmatches any digit).
The next half of the dialog box specifies how the dates will be parsed and interpreted:
- Parse as a number
- We used this option above. It basically assumes the date is a number. It may be a year or a number of days or it can be a decimal number in what ever units (positive numbers are from some time in the past, negative are from some time in the future). The suboptions here are as described above.
- Parse as calendar date
- Here we assume the date is specified in one of the standard formats which is specified in the
Date formatbox. These date formats are very flexible and can specify how to parse most date formats. Click the?button to see a description of the format. As an example,dd/MM/yyyywould denote the globally common day month year notation (MM/dd/yyyywould be the US’s confusing alternative). - Parse calendar date with variable precision
- This is parses very specific way of specifying dates which is by far the most reliable (it is used widely as the format for databases and other computer systems because of its unambiguity. This format is the equivalent of using
yyyy-MM-ddin the box above (an example date being1971-05-04). This format has an additional advantage is that the day can be ommitted (1971-05) if it is not known. Both the day and the month can be ommitted if only the year is known (1971). When using this option (as opposed to the previous one) cases where the day or day and month are ommitted are noted and the uncertainty is recorded in theprecisioncolumn. This has two benefits - firstly BEAST will place the tip in the middle of this date range (which if you don’t know when in 1971 your sequence was sampled, is probably a good choice). Secondly, it allows the uncertainty in the date to be correctly accommodated by BEAST - sampling over the date range. For more details about this, see the Tip Date Sampling how-to.
Just like the Set Dates option, if you select certain rows and use Guess Dates it will only extract dates for those taxa (you can then use a different option for others).
Importing dates from a file
The final option is the Import Dates button. This will ask you for a file which contains dates for each of the sequences. The format is simply a text file with the taxon label and the date separated by a tab character. For the RSV data set this will look like:
NYCH09@93 1993
SE05@91 1991
BE11600@94 1994
BE15471@97 1997
SE03@91 1991
MAD1@93 1993
BE1061@00 2000
.
.
.
This dialog provides the same options for specifying how the dates should be parsed as above. Any rows in the file that cannot be exactly matched to a taxon lable will be ignored and any taxa that don’t have a row in the file will not have a date specified (they will remain at zero).
References
Rambaut (2000) Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics, 16: 395-399.
Drummond, Pybus, Rambaut, Forsberg and Rodrigo (2003) Measurably evolving populations. Trends in Ecology and Evolution, 18: 481-488