Tip Date Sampling
In this tutorial, we will show various approaches to accommodate uncertainty in sampling dates using data from the West Nile virus (WNV) invasion across North America (Pybus et al., 2012). The data consists of a set of viral genome sequences which have been isolated at different points in time (heterochronous data) in different US counties. WNV is a mosquito-borne RNA virus whose primary host is birds, and was first detected in the United States in New York City in August 1999. The data are 104 genomes collected between 1999 and 2008. The taxon labels contain the sampling dates for all the sequences in the data set, as can be seen in the figure below.
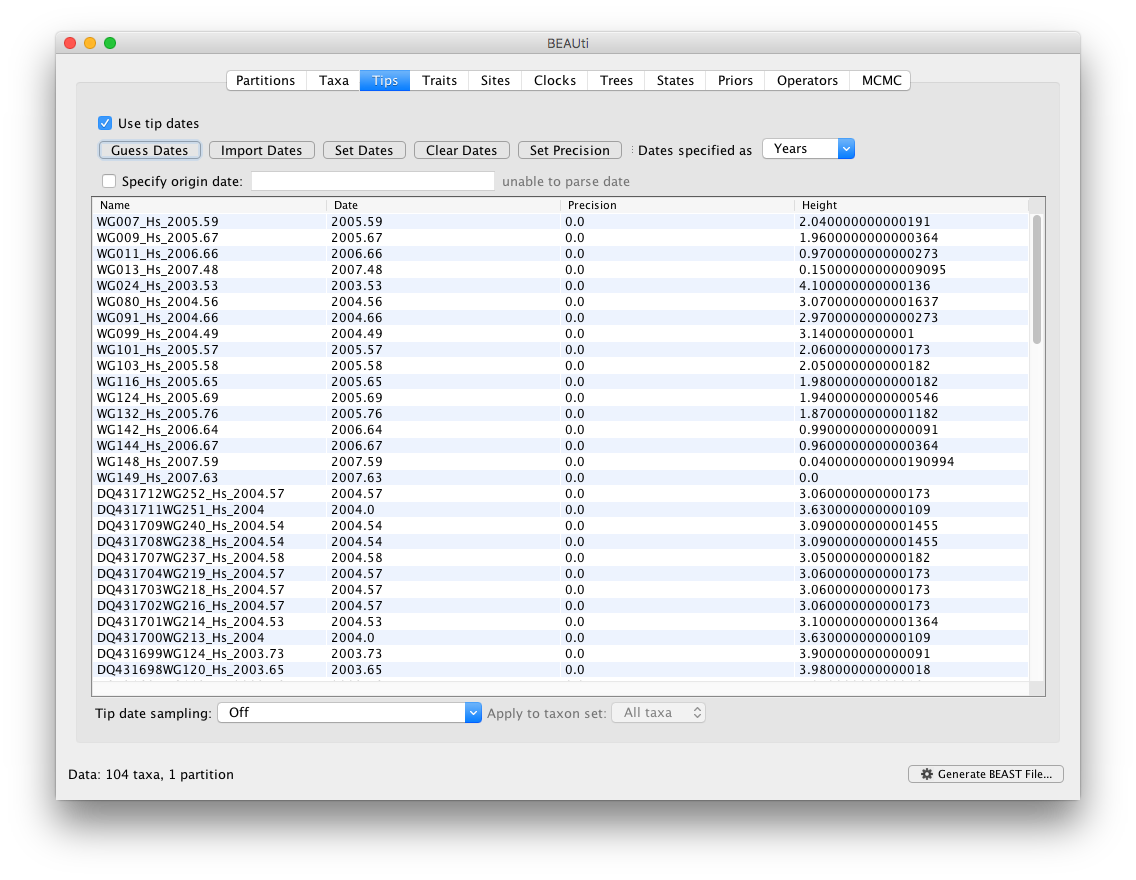
Note: To inform BEAUti/BEAST about the sampling dates of the sequences, check out the tutorial on how to extract dates from taxon labels.
Setting Precision for Selected Taxa
The ‘Height’ column lists the ages of the tips relative to time 0 (in the case of this WNV data set 2007.63). The ‘Precision’ column allows specifying with what precision the sampling times are known. This is useful in the WNV case because some sampling dates are known to the exact day, while others are only known up to the year of sampling (those without decimal in the taxa name or with the .0 decimal in the Date column), and BEAST allows to integrate over the uncertainty of the latter. To make use of the ability to adequately accommodate the uncertainty of our sampling dates, select all the taxa (holding down the cmd of shift key) with sampling dates only known up to the year in the Tips window and click on ‘Set Precision’.
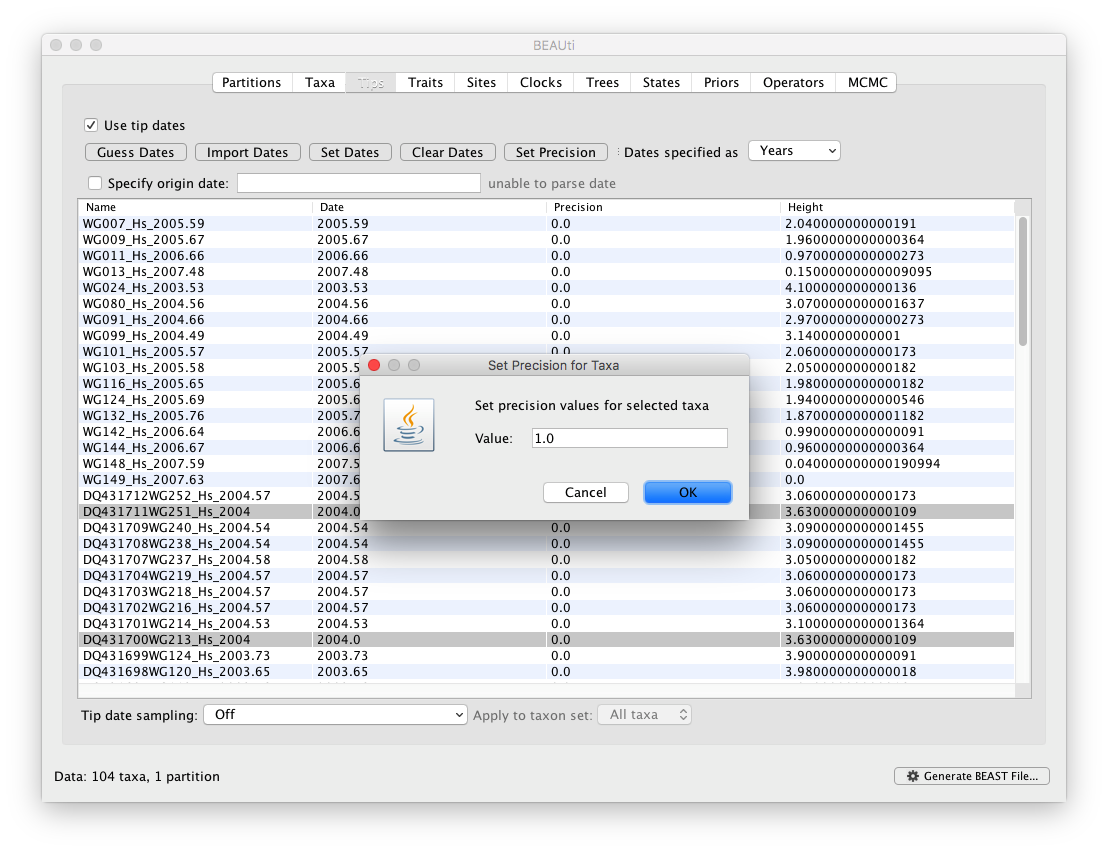
Enter (for example) ‘1.0’ as the precision value for 1 year. This will instruct BEAST to add a half year to those sampling dates (currently without visually updating the value in BEAUti) and construct a uniform window of 1 year around this new value. To estimate the respective sampling dates within the constraint of this window in the MCMC analysis, select ‘sampling uniformly from precision’ at the bottom left of the Tips panel and keep the ‘Apply to taxon set: All taxa’ default.
Setting Priors for Selected Taxa
BEAUti also provides the option to provide prior information on sampling dates. In order to enable this feature, one or more taxon sets need to be constructed.
Note: We provide a tutorial on how to create taxon sets in BEAUti.
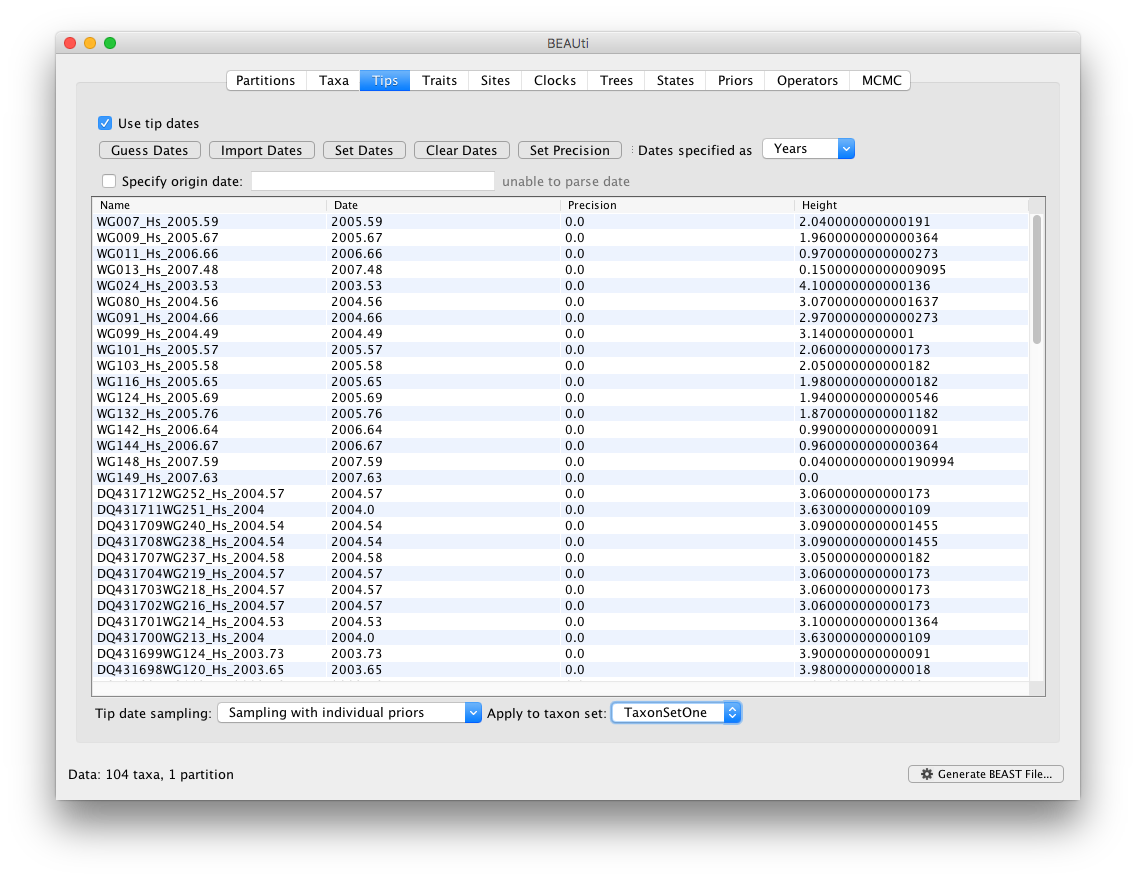
When a taxon set has been created, the option ‘Sampling with individual priors’ can be selected and applied to a taxon set of choice, here with ID ‘TaxonSetOne’.
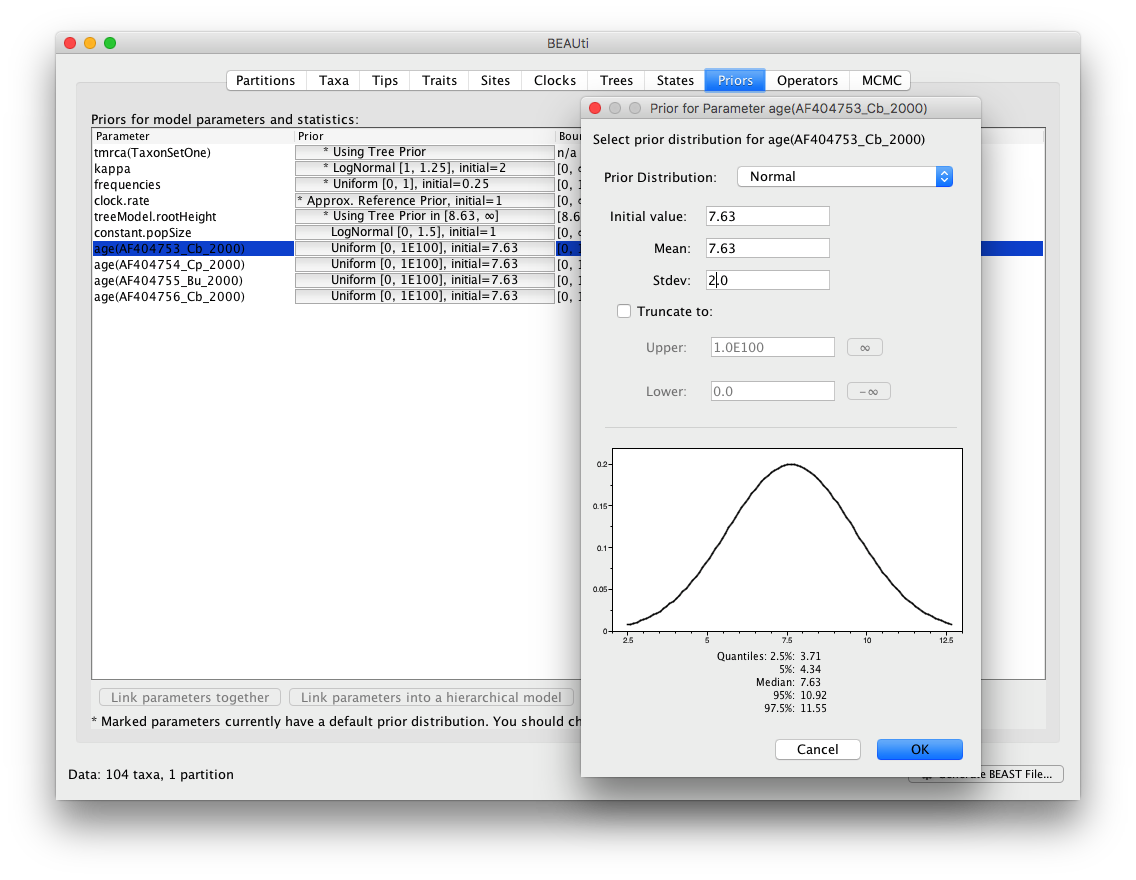
This estimation process needs a set of priors to be defined in the ‘Priors’ tab in BEAUti. Proper priors need to be defined for the node height of each taxon within the selected taxon set. Additionally, it’s possible to define a prior on the time to most recent common ancestor (TMRCA) of the selected taxon set.
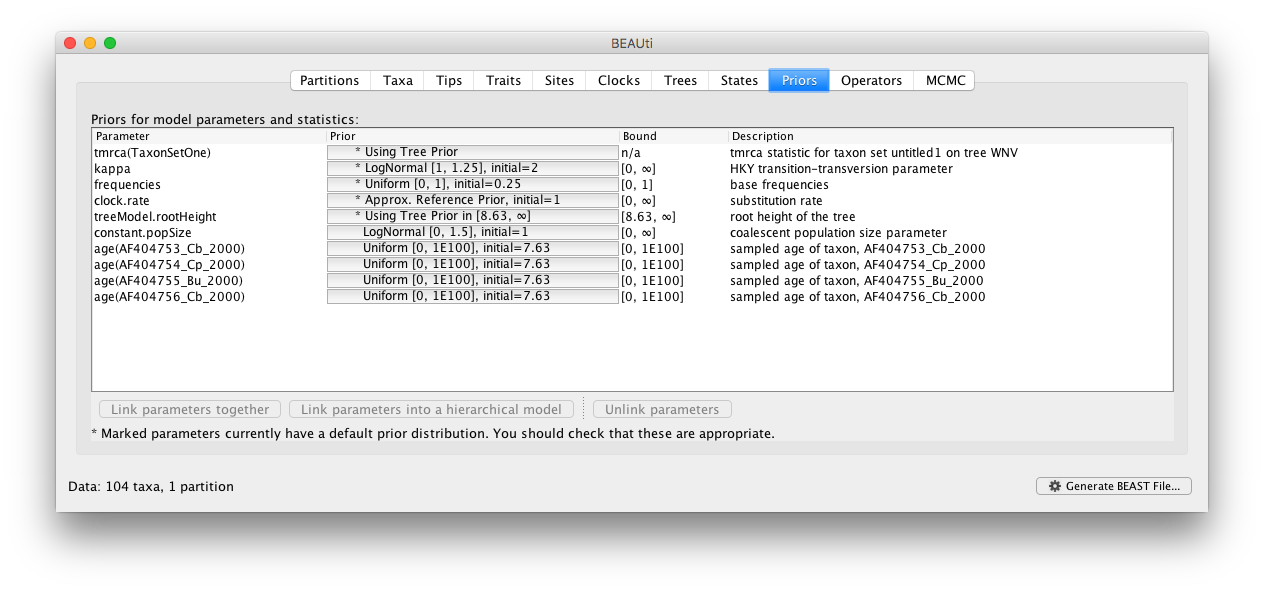
To alter the default prior on the node height of (for example) taxon ‘AF404753_Cb_2000’, click on the current prior choice ‘Uniform[0, 1E100], initial=7.63’. Here, it’s shown how to put a Normal prior with mean equal to 7.63 and standard deviation equal to 2.0 on the node height.
References
Pybus et al. (2012) Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc. Natl. Acad. Sci. USA 109(37): 15066-15071.